2018.03.04.3
Files > Volume 3 > Vol 3 No 4 2018
INVESTIGATION / RESEARCH
Gene expression profile in cervical carcinoma cells treated with HeberFERON
Perfil de expresión génica en células de carcinoma cervical tratadas con HeberFERON
Dania Vázquez-Blomquist1*, Julio Raúl Fernández1, Jamilet Miranda2, Isabel Guillén1, Seidy Pedroso1, Alexander Martín2, María Elena Ochagavia2, José Angel Silva3, Regla Estrada3, Omar Gell3, Lidia Inés Novoa1, Daniel Palenzuela1, Iraldo Bello4.
Available from: http://dx.doi.org/10.21931/RB/2018.03.04.3
ABSTRACT
Interferon-alpha (IFN-α) and gamma (IFN-γ) are important cytokines with multiple functions. HeberFERON is a co-formulation of recombinant IFN-α2b and IFN-γ that shows improved pharmacodynamics properties and stronger antitumor response than individual IFNs. The aim of this study was to investigate the differentially expressed genes by HeberFERON in relation to their IFN components by a Suppressive Subtractive Hybridization (SSH) study. Two subtractive cDNA libraries were constructed from HEp-2 cells independently treated with recombinant IFN-α2b and IFN-γ for 72 hours as tester, and cells treated with HeberFERON as driver and vice versa. Near 300 cloned PCR products were sequenced and compared to the database in GenBank and BLAST. We obtained homology to 36 known proteins coding genes. Genes for ribosomal proteins and translation factors besides rRNAs 18S and 28S, cytoskeleton related proteins and proteins participating in antigen presentation and immune responses were mainly identified, using DAVID and GeneCodis tools. Validation of differential gene expression (p< 0.05) of genes from the main biological processes components by quantitative PCR (qPCR) showed diverse gene signature by individual IFNs or HeberFERON.
Keywords: gene expression, HEp-2, interferon alpha, interferon gamma, quantitative PCR, suppressive subtractive hybridization
RESUMEN
El interferón-alfa (IFN-α) y el gamma (IFN-γ) son importantes citoquinas con múltiples funciones. HeberFERON es una co-formulación de IFN-α2b recombinante e IFN-γ que muestra propiedades farmacodinámicas mejoradas y una respuesta antitumoral más fuerte que los IFN individuales. El objetivo de este estudio fue investigar los genes expresados diferencialmente por HeberFERON en relación con sus componentes de IFN mediante un estudio de Hibridación Subtractiva Supresiva (SSH). Se construyeron dos bibliotecas de ADNc sustractivas a partir de células HEp-2 tratadas independientemente con IFN-α2b recombinante e IFN-γ durante 72 horas, y las células tratadas con HeberFERON como conductor y viceversa. Cerca de 300 productos de PCR clonados fueron secuenciados y comparados con la base de datos en GenBank y BLAST. Obtuvimos homología con 36 proteínas conocidas que codifican genes. Se identificaron principalmente genes para proteínas ribosómicas y factores de traducción además de los ARNr 18S y 28S, proteínas relacionadas con citoesqueleto y proteínas que participan en la presentación de antígenos y respuestas inmunitarias, utilizando las herramientas DAVID y GeneCodis. La validación de la expresión génica diferencial (p <0.05) de los genes de los principales componentes de los procesos biológicos mediante PCR cuantitativo (qPCR) mostró una firma de genes diversa por IFN individuales o HeberFERON.
Palabras clave: expresión génica, HEp-2, interferón alfa, interferón gamma, PCR cuantitativa, hibridación sustractiva supresiva
INTRODUCTION
Interferons (IFN-α, -β, -λ and -γ) are a multigene family of cytokines that possess a wide range of biological functions including antiviral, anti-proliferative, pro-apoptotic, anti-angiogenesis, anti-fibrotic, neuromodulators and other effects 1, 2, through activation of related pathways 3. These pathways involve specific IFN type I and type II receptors, which initiate activation through JAK-STAT cascades. Type I IFNs interact with the IFNα/β receptor (IFNAR) subunits composed by IFNAR1 and IFNAR2 associated with tyrosine kinase 2 (TYK2) and Janus kinase 1 (JAK1); while IFN-γ binds to the IFN-γ receptor (IFNGR) receptor subunits composed by IFNGR1 and IFNGR2 associated with JAK1 and JAK2 4.
Thus, IFNs induce the expression of hundreds of IFN-regulated genes (IRGs) via the JAK-STAT pathway 5. Some of IRGs are regulated by both types of IFNs, whereas others are selectively induced by distinct IFNs through drastic changes in genomic binding locations in a manner dependent on the combinational involvement of STAT1 and STAT2 6. Depending of doses, treatment time and other factors, IFN-α and IFN-γ signaling may interfere or potentiate each other 7. Contemporary studies as ChIP-chip analysis of STAT1 and STAT2 targets coupled to quantitative gene-specific PCR (ChIP-qPCR), screening of DNA microarrays or tiling arrays (ChIP-chip), or high-throughput DNA sequencing (ChIP-seq) methods have permitted a more comprehensible picture of how the complex machinery composed by transcription factors, transcriptional co-regulators, histone modifiers, and other players is working for the regulation of IFN target genes 4.
HeberFERON is a co-formulation of IFN-α2b and IFN-γ, with improved pharmacodynamics properties 8 that has demonstrated better results than the individual IFNs in the treatment of basocellular and spinocellular carcinomas 9, 10. In an attempt to evaluate the gene expression pattern promoted by HeberFERON and potential distinctive regulation of the combination with respect to separated IFNs, we performed a SSH experiment 11 linked to qPCR in HEp-2 cell line, representing cervical carcinoma tissue. The understanding of the biological effects that distinguish HeberFERON from their individual components will aid to the optimal clinical application of this formulation in the future.
MATERIALS AND METHODS
Biological Reagents. Recombinant (r) interferons, rIFN-a2b and rIFN-g, and the pharmaceutical co-formulation of both rIFN-a2b and rIFN-g, HeberFERON, were produced at CIGB, Havana, Cuba.
Cell treatment for suppression subtractive hybridization (SSH) experiment.
HEp-2 (ATCC-CCL23) cell line, (human cervix carcinoma), was grown in MEM-CANE (Gibco, USA) containing gentamycin (50 mg/mL) (Gibco, USA) and 10% fetal bovine serum (FBS) (Gibco, USA). Cells were seeded into 75cm2 dishes at 3-4 x104 cells/ mL in culture medium containing 10% FBS, incubated at 37 oC and 5% of CO2. Twenty four hours later, the medium of the treatment groups were refreshed with rIFN-α2b (75 IU/ml), rIFN-g (5 IU/ml) or HeberFERON while the control cells received only cell culture medium with antibiotic and serum. Cells were incubated for another 72h.
Construction of SSH library.
Total RNA was extracted from cells by TriReagent procedure (Sigma, USA) and DNase I treatment (Promega, USA). First-strand cDNA synthesis was carried out following SuperScript II reverse transcriptase Invitrogen kit instructions (Invitrogen, USA) from 5μg of total RNA. The second strand cDNA synthesis (dscDNA) was carried out from the first strand and a mixture with DNA polymerase I, RNase H and dNTPs (Promega, USA) at 14 ºC overnight. The dscDNA from HeberFERON -treated (population 1) and [IFN-α2b + IFN-g]-treated (population 2) HEp-2 cells were used as tester and driver, respectively, in a first hybridization (SSH1) and they were exchanged in a second hybridization (SSH2) to obtain both genes upregulated and downregulated. Subtracted cDNA libraries were constructed using Clontech PCR-Select cDNA subtraction kit (Clontech Laboratories, USA), following the manufacturers protocol. The subtracted tester dscDNA was amplified in suppression and nested PCR to enrich only the “differential population”. The nested PCR products from libraries SSH1 and SSH2 were ligated into pGEM T-Easy Vector (Promega, USA), transformed into DH10B E. coli cells and screened on LB plates containing ampicillin/X-gal/IPTG at standard concentrations. More than 3000 white colonies were obtained. Plasmids from selected clones were purified using a Qiagen plasmid mini kit (Qiagen, USA).
EST sequencing and bioinformatics analysis.
About 300 clones were sequenced using an automated sequencer (Macrogen, Korea) and submitted to GenBank for homology analysis. Nucleic acid homology searches were performed using the BLAST program (National Institutes of Health, Bethesda, Md.). DAVID and GeneCodis were used for Gene Annotation and Functional clustering analysis 12, 13. Web sites for IFNs, INTERFEROME V1.0 and V2.01 were consulted to find genes described as IRGs in our list (http://interferome.its.monash.edu.au/interferome/home.jspx and https://interferome-v1.erc.monash.edu.au) (October 22, 2018) 14, 15.
Quantitative PCR validation.
We validated gene expression differences among treatment conditions for a subset of transcripts derived from SSH by qPCR as described 16. The design included two biological replicates of untreated, IFN-a2b- treated, IFN-g- treated and HeberFERON - treated cell samples; two replicates of cDNA reactions (from 1µg of total RNA) from each and three technical replicates. As a result, we had 12 data per sample per gene. Primers are listed in Table 1S (Supplemental Materials). Statistically significant results were considered for p<0.05 after reference gene normalization.
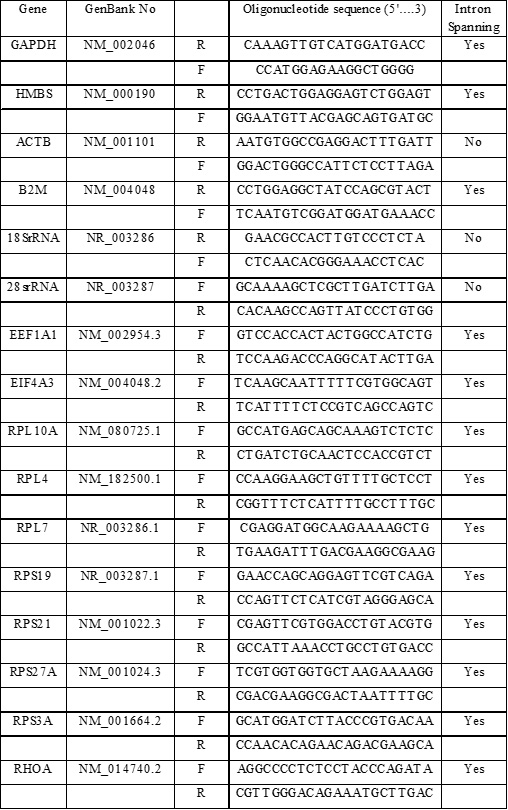
Table 1S. Genes evaluated and Primers information. A summary of the Gene symbols and GenBank Number (No), Sequences for both oligonucleotides (5´…3´; Forward:F and Reverse: R) and intron spanning characteristic are provided.
RESULTS
Identification of differentially expressed genes after HeberFERON treatment in HEp-2 cells
From 288 clones sequenced, 215 clones were found to be highly homologous (92%-100%, E value near 0) with 36 known genes. High numbers of hits were obtained for 18S and 28S ribosomal RNAs (rRNA); four genes had more than 10 hits (RHOA, RPS21, C19orf42, and RAB7L1). Other 30 genes were also identified (Table 1).

Table 1. Homology analysis results of positive clones with Gen-Bank database. GenBank Accession Number, Gene Description, Gene ID and Name are provided. Number (No) of hits for each gene is also included.
Using DAVID and GeneCodis we annotated genes in relation to biological process, molecular function and cell compartment and obtained functional clusters (Table 2). The identified genes code for: structural proteins constituent of ribosome (RPL4, 7, 10A, 24, RPS3A, 16, 19, 21 and 27A); proteins participating in protein synthesis (EIF4A3, EEF1A1), in regulation of actin cytoskeleton (ACTG1, ACTB, RHOA) and in antigen processing and presentation and immune response (B2M, HLA-C, HLA-B, HSPD1, HSPA5, HSP90AB1).

TABLE 2A

TABLE 2B
Tabla 2. Gene Annotation Clustering by DAVID (A) and GeneCodis (B). (A) Each annotation cluster has an Enrichment Score associated, a Category and Term from the Databases consulted, a list of Genes included in categories and the p value associated. (B) Each Biological Process included a Number and list of Genes. P values were obtained through Hypergeometric analysis corrected by FDR method, showing the significance of each process (Hyp_c).
Cross-referencing the gene list (Table 1) with the INTERFEROME V1.0 and V2.01 databases 14, 15 highlighted C2orf50 gene as the only one that had not been reported as IFN response gene in humans.
We selected transcripts from coding genes participating in the main biological processes for validation by qPCR in the HEp-2 cells untreated or treated with individual IFNs or HeberFERON. As described before 16, GAPDH and HMBS genes were the least variable and were used for qPCR normalization. Table 3 shows the factor of change for each gene in each experimental condition (treatment with IFN-α2b, IFN-γ or HeberFERON) respect to the untreated control after normalization with the two reference genes.
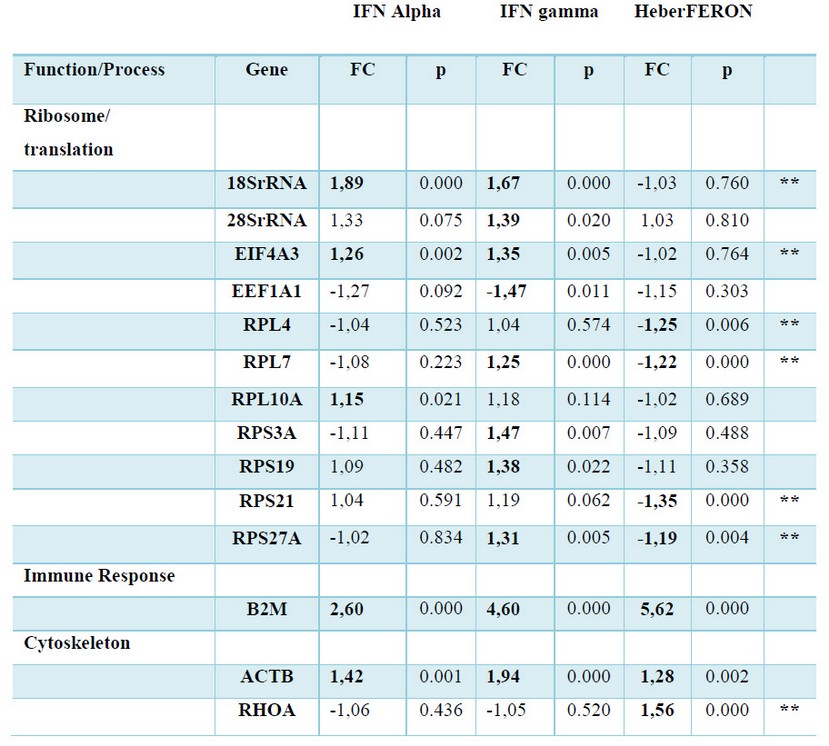
Table 3 Gene expression (mRNA) following treatment with IFNα (Alpha), IFNγ (gamma) and the combination HeberFERON. Factor of Change (FC) respect to untreated cells, calculated by REST 2009, and the p values associated to each comparison are shown. In Bold and Shadows formats we show genes which have statistically significant differences (p < 0.05). Genes are grouped by Function and Process. Double asterisks pointed to the genes showing new regulation pattern of HeberFERON respect to IFNα (Alpha) and IFNγ (gamma).
Using RT-qPCR fourteen genes were validated as HeberFERON response genes (Table 3). B2M and ACTB genes were up-regulated by IFN-α2b, IFN-γ and HeberFERON, confirming they are IRGs as it has been reported before 5, 17-19.
In the cases of a group of 18SrRNA, 28SrRNA, EIF4A3, EEF1A1, RPL10A, RPS3A, and RPS19 genes, non-static significant differences were detect for HeberFERON gene expression regulation (see Table 3). Conversely, both IFNs up-regulated 18SrRNA and EIF4A3 gene expressions; while RPL10A, 28SrRNA, RPS3A and RPS19 genes were upregulated solely by IFN-α2b or IFN-γ. EEF1A1 gene was the only gene down-regulated by the treatment with IFN-γ. This gene regulation behavior is an evidence of a differential gene expression pattern of HeberFERON with respect to separated IFNs. Another differential gene signature was detected for RPL4 and RPS21 or RHOA genes that were solely down-regulated or up-regulated, respectively, by HeberFERON. A more intriguing gene regulation pattern was observed for RPL7 and RPS27A genes. In both cases, the separated IFNs (IFN-α2b, no effect; IFN-γ up-regulation) and HeberFERON (down-regulation) promoted different regulation pattern.
The carefully observation of these transcriptional signatures identified potential antagonistic or synergistic effect of HeberFERON.
Antagonism between IFN-α2b and IFN-γ is expected for those genes where each IFN regulates the gene differently. These are the cases of: 28SrRNA, EEF1A1, RPL7, RPL10A, RPS3A, RPS19 and RPS27A genes. Another kind of antagonist was observed where both IFNs up-regulated the gene expression but HeberFERON, unexpectedly, had no effect on the mRNA expression of these genes. We have the cases of 18SrRNA and EIF4A3F as examples. Additionally, a clear antagonist effect between IFNα-2b and IFN-γ is observed for the regulation of RPL4 and RPS21 genes, where separately IFNs had no effect on regulation, while HeberFERON down-regulated the expression of both genes.
Additive or synergistic effect of the combination of both IFNs could be the cause of the significant increase in gene expression of RHOA gene by HeberFERON.
DISCUSSION
A subtractive hybridization assays experiment was carried out in HEp-2 cell line with sensitivity to growth arrest by IFN-g 20, IFN-α 21 and their co-formulation as HeberFERON 10. In this study, we used this cell line as a model to firstly understand what distinguish HeberFERON from individual IFNs actions, at the transcript level.
As it has been reported in previous microarrays studies 5, 17-19 and compiled in IFN Databases INTERFEROME V1.0 14, 22 and V2.01 15, we identified HeberFERON differentially expressed genes encoding proteins that participate in Antigen processing and presentation and Immune Response (B2M, HLA-C, HLA-B, HSPD1, HSPA5, HSP90AB1), Cytoskeleton regulation (ACTB, ACTG1, RHOA) and a high proportion in protein translation (RPL10A, RPL24, RPL4, RPL7, RPS16, RPS19, RPS21, RPS27A, RPS3A, EEF1A1 and EIF4A3). High percentage of hits was for rRNAs 18S and 28S.
IFN Database INTERFEROME V1.0 14, 22 shows 69% of the IFN-g–regulated genes are also induced by type I IFNs. In comparison to type I IFNs used alone, the addition of type II IFN caused enhanced expression not only of many of the genes correlated with the direct antiviral state but also of genes involved in Antigen Presentation to cytotoxic T lymphocytes (CTLs) and Apoptosis 22.
SSH confirmed 36 differentially expressed genes from 215 sequenced clones, representing a 17%, in consistence with previous publications where the number of genes obtained by SSH represents less than 25% of the number of sequenced clones 23, 24.
The differences in gene regulation after 72 hour of incubation time with IFNs could explain, at least in part, the differential gene expression patterns detected. A recent study examining gene expression in human cervical cancer cell line found that although IFN-α and IFN-β induced comparable levels of transcription at early time points, IFN-α induced transcription declined after 8 hours 25. This decrease was associated with the expression of the IFN stimulated gene USP18 (UBP43), which interacts with the IFNAR2 and inhibits signaling through JAK1 26. Moreover, treatment with IFN-γ for 72 hours markedly inhibits IFN-α-activated STAT1, STAT2 and STAT3; whereas a 24 hours’ treatment with IFN-γ slightly enhanced IFN-α-activated STAT1 4.
HeberFERON antagonizes the effect of IFN-α2b and IFN-γ on the expression of 18SrRNA, EIF4A3 (helicase that promotes tumorigenesis 27), RPL4 (inhibitor of normal physiological levels of p53 28), RPL7 (associated with an increased risk factor at early stages of colon recto carcinoma development 29), RPS27A (promotor of proliferation, cell cycle progression and inhibitor of apoptosis in solid tumors, advanced-phase chronic myeloid leukemia (CML) and acute leukemia (AL) patients 30, 31), and RPS21 (its reduction is coupled to antitumor effect of ruthenium compound 32). Anti-cancer effect of HeberFERON at blocking translation would be more effective when multiple intervening factors can be inhibited in combination.
Antagonism between IFN-α and IFN-γ has been reported by several authors. The antagonism could involve regulation of IFN receptor expression, as observed by Rayamajhi et al, when IFN type I reduced the expression of IFNGR1 in macrophages infected with L. Monocytogenes, with the corresponding suppression of host responsiveness to IFN-γ 33. In macrophages, interferon consensus sequence binding protein (ICSBP) mRNA and protein are strongly induced by IFN-γ, but only marginally by IFN type I. When both IFNs are present, IFN type I antagonizes IFN-γ-induced ICSBP mRNA and protein synthesis 34.
Regulation of phosphorylation of transcriptional factors involved in IFN type I and type II signaling could also explain the antagonism observed for HeberFERON with respect to separated IFNs. For example, overexpression of protein tyrosine phosphatase Shp1 in endothelial cells abrogated IFN type I signaling through a GAS site, suggesting a role of level of Shp1 on the interference between IFN types I and II signaling pathways 35.
The increased gene expression of RHOA stimulated by HeberFERON could indicate an additive or synergic effect. IFN-α 36 and IFN-γ 37 have been involved in the reorganization of the cell cytoskeleton through RHOA, with impact in the cell growth. The upregulation of RHOA by HeberFERON could be beneficiated from the described crosstalk between both type of IFNs 38 and the further regulated expression of STAT1 via c-Jun-mediated production of basal levels of IFN-β 39. In this context, we could remark the facts that STAT1 and the stimulation of c-Jun expression could be involved in the regulation of RHOA gene expression 39, 40.
The diverse mode of gene regulation revealed in this work by the combination of IFN-α2b and IFN-γ (HeberFERON), is congruent with the recent study of the ENCODE project performed on genomic binding sites suggesting that transcription factors often show different co-association patterns in binding sites, and the binding of one transcriptional factor affects the preferred binding partners of others 41. Furthermore, efficient transcriptional activation of STAT1 target genes requires posttranslational modification of STAT1 and the recruitment of coactivators and histone and chromatin modifying complexes 4.
As part of this work we obtained the gene C2orf50 regulated by individual IFNs or their combination that was not previously described as IRGs in humans 14, 15. This gene is poorly characterized [https://www.uniprot.org/uniprot/Q96LR7] and the understanding of their participation in the mechanism of action of HeberFERON could be an interesting point in the future.
In spite of our study examined only a small number of IRGs, it suggests that compared with transcriptional patterns of separated IFNs, HeberFERON induces a unique transcriptional signature after 72 hour of cell treatment. The meaning of this new signature should be taken into account for clinical translation.
Competing interests and Funding
The authors declare that they have no competing interests. All Authors are (were) employees of the Center for Genetic Engineering and Biotechnology (CIGB), Havana where IFNa2b, IFNg and HeberFERON are produced. The study was financed by CIGB.
ACKNOWLEDGEMENTS
We would like to thank Amanda Colarte, Adelaida Villarreal and Tamara Díaz for their technical support.
REFERENCES
1. Pestka S. The Interferons: 50 Years after Their Discovery, there is much more to learn. J Biol Chem. 2007 Jul 13;282(28): 20047-51. DOI: 10.1074/jbc.R700004200.
2. Bekisz J, Baron S, Balinsky C, Morrow A, Zoon KC. Antiproliferative properties of Type I and Type II interferon. Pharmaceuticals (Basel). 2010 Mar 30;3(4): 994-1015. DOI: 10.3390/ph3040994.
3. Katsoulidis E, Kaur S, Platanias LC. Deregulation of interferon signaling in malignant cells. Pharmaceuticals (Basel). 2010 Feb 4;3(2):406-418. DOI: 10.3390/ph3020406.
4. Satoh J, Tabunoki H. Comprehensive Profile of ChIP-Seq-based STAT1 Target genes suggests the complexity of STAT1-mediated gene regulatory mechanisms. Gene Regul Syst Bio. 2013 Mar 26;7: 41-56. DOI: 10.4137/GRSB.S11433.
5. Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes dif-ferentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998 Dec 22;95(26): 15623-8.
6. Hartman SE, Bertone P, Nath AK, Royce TE, Gerstein M, Weissman S et al. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 2005 Dec 15;19(24): 2953-68. DOI: 10.1101/gad.1371305.
7. Radaeva S, Jaruga B, Kim WH, Heller T, Liang TJ, Gao B. Interferon-γ inhibits interferon-α signaling in hepatic cells: evidence for the involvement of STAT1 induction and hyperexpression of STAT1 in chronic hepatitis C. Biochemical Journal 2004 Apr 01;379(1): 199-208. DOI: 10.1042/bj20031495.
8. García-Vega Y, García-García I, Collazo-Caballero SE, Santely-Pravia EE, Cruz-Ramírez A, Tuero-Iglesias AD et al. Pharmacokinetic and pharmacodynamic characterization of a new formulation containing synergistic proportions of interferons alpha-2b and gamma (HeberPAG®) in patients with mycosis fungoides: an open-label trial. BMC Pharmacology and Toxicology 2012,13: 20. DOI: 10.1186/2050-6511-13-20.
9. Anasagasti-Angulo L, García-Vega Y, Barcelona-Perez S, López-Saura P, Bello-Rivero I. Treatment of advanced, recurrent, resistant to previous treatments basal and squamous cell skin carcinomas with a synergistic formulation of interferons. Open, prospective study, BMC Cancer 2009, 9:262 DOI: 10.1186/1471-2407-9-262.
10. Bello-Rivero I, Garcia-Vega Y, Valenzuela-Silva C, Bello-Alvarez C, Vázquez-Blomquist D, Lopez-Saura P. Development of a new formulation of interferons (HEBERPAG) for BCC treatment. J Cancer Res Ther 2013;1(10): 235-243. DOI: 10.14312/2052-4994.2013-36.
11. L. Diatchenko YF, Laut C, Campbell AP, Chenchik A, Moqadam F, Huang B et al. Suppression subtractive hybridization: A method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996 Jun 11;93(12): 6025-30.
12. Dennis GJr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biology 2003,4: R60.
13. Tabas-Madrid D, Nogales-Cadenas R, Pascual-Montano A. GeneCodis3: a non-redundant and modular enrichment analysis tool for functional genomics. Nucleic Acids Res. 2012 Jul;40(Web Server issue):W478-83. DOI: 10.1093/nar/gks402.
14. Samarajiwa SA, Forster S, Auchettl K, Hertzog PJ. INTERFEROME: the database of interferon regulated genes. Nucleic Acids Res. 2009 Jan;37(Database issue): D852-7. DOI: 10.1093/nar/gkn732.
15. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H et al. INTERFEROME v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 2013 Jan;41(Database issue): D1040-6. DOI: 10.1093/nar/gks1215.
16. Vázquez-Blomquist D, Fernández JR, Miranda J, Bello C, Silva JA, Estrada RC et al. Selection of reference genes for use in quantitative reverse transcription PCR assays when using interferons in U87MG. Mol Biol Rep. 2012 Dec;39(12): 11167-75. DOI: 10.1007/s11033-012-2026-9.
17. de Veer MJ, Holko M, Frevel M, Walker E, Der S, Paranjape JM, Silverman RH et al. Functional classification of interferon-stimulated genes identified using microarrays J Leukoc Biol. 2001 Jun;69(6): 912-20. DOI: 10.1189/jlb.69.6.912.
18. Tan H, Derrick J, Hong J, Sanda C, Grosse WM, Edenberg HJ et al. Global transcriptional profiling demonstrates the combination of Type I and Type II interferon enhances antiviral and immune responses at clinically relevant doses. J Interferon Cytokine Res. 2005 Oct;25(10): 632-49. DOI: 10.1089/jir.2005.25.632.
19. Sanda C, Weitzel P, Tsukahara T, Schaley J, Edenberg HJ, Stephens MA et al. Differential Gene Induction by Type I and Type II Interferons and Their Combination. J Interferon Cytokine Res. 2006 Jul;26(7): 462-72. DOI: 10.1089/jir.2006.26.462.
20. de la Maza LM, Peterson EM. Dependence of the in vitro antiproliferative activity of recombinant human -interferon on the concentration of tryptophan in culture media. Cancer Res. 1988 Jan 15;48(2): 346-50.
21. Danielescu G, Maniu H, Oprescu E, Jucu V, Georgescu T, Cajal N. Anti-proliferative and antiviral effects of human alpha-interferon on tumor cells. Virologie. 1987 Apr-Jun;38(2): 83-93.
22. Hertzog P, Forster S, Samarajiwa S. Systems Biology of Interferon Responses. J Interferon Cytokine Res. 2011 Jan;31(1): 5-11. DOI: 10.1089/jir.2010.0126.
23. Patzwahl R, Meier V, Ramadori G, Mihm S. Enhanced expression of interferon-regulated genes in the liver of patients with chronic Hepatitis C Virus infection: detection by suppression-subtractive hybridization. J Virol. 2001 Feb;75(3): 1332-8. DOI: 10.1128/JVI.75.3.1332-1338.2001.
24. Qu JH, Cheng J, Zhang LX, Zhang LY, Zhong YW, Liu Y et al. Identification of genes upregulated by recombinant interferon-alpha in HepG2 cells by suppressive subtractive hybridization analysis. Hepatobiliary Pancreat Dis Int. 2007 Jun;6(3): 290-3.
25. Francois-Newton V, Livingstone M, Payelle-Brogard B, Uz_e G, Pellegrini S. USP18 establishes the transcriptional and anti-proliferative interferon a/b differential. Biochem J. 2012 Sep 15;446(3): 509-16. DOI: 10.1042/BJ20120541.
26. Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KGS, Fuchs SY et al. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. The EMBO Journal 2006; 25: 2358-2367. DOI: 10.1038/sj.emboj.7601149.
27. Rubio CA, Weisburd B, Holderfield M, Arias C, Fang E, DeRisi JL et al. Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 2014;15(10): 476. DOI: 10.1186/s13059-014-0476-1.
28. He X, Li Y, Da MS, Sun XX. Ribosomal protein L4 is a novel regulator of the MDM2-p53 loop. Oncotarget. 2016 Mar 29;7(13): 16217-26. DOI: 10.18632/oncotarget.7479.
29. Boleij A, Roelofs R, Schaeps RMJ, Schülin T, Glaser P, Swinkels DW et al. Increased exposure to bacterial antigen RpL7/L12 in early stage colorectal cancer patients. Cancer. 2010 Sep 1;116(17): 4014-22. DOI: 10.1002/cncr.25212.
30. Wang H, Xie B, Kong Y, Tao Y, Yang G, Gao M et al. Overexpression of RPS27a contributes to enhanced chemoresistance of CML cells to imatinib by the transactivated STAT3 . Oncotarget. 2016 Apr 5;7(14): 18638-50. DOI: 10.18632/oncotarget.7888.
31. Wang H, Yu J, Zhang L, Xiong Y, Chen S, Xing H et al. RPS27a promotes proliferation, regulates cell cycle progression and inhibits apoptosis of leukemia cells. Biochem Biophys Res Commun. 2014 Apr 18;446(4): 1204-10. DOI: 10.1016/j.bbrc.2014.03.086
32. Elumalai P, Jeong YJ, Park DW, Kim DH, Kim H, Kang SC et al. Antitumor and biological investigation of doubly cyclometalated ruthenium (II) organometallics derived from benzimidazolyl derivatives. Dalton Trans. 2016 Apr 21;45(15): 6667-73. DOI: 10.1039/c5dt04400f.
33. Rayamajhi M, Humann J, Penheiter K, Andreasen K, Lenz LL. Induction of IFNα/β enables Listeria monocytogenes to suppress macrophage activation by IFNg. J Exp Med. 2010 Feb 15;207(2): 327-37. DOI: 10.1084/jem.20091746.
34. Fultz MJ, Vogel SN. Analysis of the antagonist effect of IFN-alpha on IFN-gamma-induced interferon consensus sequence binding protein messenger RNA in murine macrophages. J Inflamm. 1998;48(1): 28-39.
35. Min W, Pober JS, Johnson DR. Interferon induction of TAP1: the phosphatase SHP-1 regulates crossover between the IFN-alpha/beta and the IFN-gamma signal-transduction pathways. Circ Res. 1998 Oct 19;83(8): 815-23.
36. Yibing X, Jianwu L, Ferguson GD, Mercurio F, Khambatta G, Morrison L et al. Immunomodulatory drugs reorganize cytoskeleton by modulating Rho GTPases, Blood. 2009 Jul 9;114(2): 338-45. DOI: 10.1182/blood-2009-02-200543.
37. Utech M, Ivanov AI, Samarin SN, Bruewer M, Turner JR, Mrsny RJ et al. Mechanism of IFN-γ-induced endocytosis of tight junction proteins: Myosin II-dependent vacuolarization of the apical plasma membrane. Mol Biol Cell. 2005 Oct;16(10): 5040-52. DOI: 10.1091/mbc.e05-03-0193.
38. Gough DJ, Messina NL, Hii L, Gould JA, Sabapathy K, Robertson AP et al. Functional Crosstalk between Type I and II Interferon through the regulated expression of STAT1. PLoS Biol. 2010 Apr 27;8(4): e1000361. DOI: 10.1371/journal.pbio.1000361.
39. Wang S, Koromilas AE. Stat1 is an inhibitor of Ras-MAPK signaling and Rho small GTPase expression with implications in the transcriptional signature of Ras transformed cells. Cell Cycle. 2009 Jul 1;8(13): 2070-9. DOI: 10.4161/cc.8.13.8891.
40. Marinissen MJ, Chiariello M, Tanos T, Bernard O, Narumiya S, Gutkind JS. The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol Cell. 2004 Apr 9;14(1): 29-41. DOI: 10.1016/S1097-2765(04)00153-4.
41. Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012 Sep 6;489(7414): 91-100. DOI: 10.1038/nature11245.
Recibido:15 october 2018
Aprobado: 17 november 2018
Dania Vázquez-Blomquist PhD1*, Julio Raúl Fernández PhD1, Jamilet Miranda MSc2, Isabel Guillén PhD1, Seidy Pedroso PhD1, Alexander Martín MSc2, María Elena Ochagavia MSc2, José Angel Silva MSc3, Regla Estrada3, Omar Gell3, Lidia Inés Novoa PhD1, Daniel Palenzuela MSc1, Iraldo Bello PhD4.
Affiliation for all authors: Department of: 1System Biology, 2Bioinformatics, 3Oligonucleotides Synthesis; 4Clinical Assays Area. Center for Genetic Engineering and Biotechnology. Ave. 31 e/158&190, Playa, 10600, Havana, Cuba.
*Corresponding Author:
Telephone: (53) 7208-04-28 extension 146/ Fax: (53) 7250-44-94
