2023.08.03.21
Files > Volume 8 > Vol 8 No 3 2023
Cloning, expression, purification and characterization of the recombinant nucleocapsid protein from SARS-CoV-2 and its combination with a CpG ODN-39M
Yadira Lobaina1,3†, Alexis Musacchio1,2,3†, Panchao Ai1,5, Rong Chen1,5, Edith Suzarte2, Changyuan Tan1,5, Ricardo Silva4, Ke Yang1,5*, Yasser Perera1,2,3* and Lisset Hermida1,3,4*
and Lisset Hermida1,3,4*
1 Research Department, China-Cuba Biotechnology Joint Innovation Center (CCBJIC) Lengshuitan District, Yongzhou City, Hunan, 425000, China
2 Research Department, Center for Genetic Engineering and Biotechnology, Havana 10600, Cuba
3 Yongzhou Zhong Gu Biotechnology Co., Ltd, Yangjiaqiao Street, Lengshuitan District, Yongzhou City, Hunan, 425000, China
4 Representative Office BCF in China. Jingtai Tower, No. 24 Jianguomen Wai Street. Chaoyang District, Beijing, PC 100022, China
5 Yongzhou Development and Construction Investment Co.Ltd. (YDCI), Changfeng Industry Park, Yongzhou Economic and Technological Development Zone, No. 1 Liebao Road, Lengshuitan District, Yongzhou City, Hunan Province, China
* Correspondence: KY [email protected], +86 13548555536, YP [email protected] +86 16681479525 DOI: and LH [email protected], +86 18510713630 DOI:
Both first authors contributed equally to the work.
Available from: http://dx.doi.org/10.21931/RB/2023.08.03.21
ABSTRACT
Several studies have reported the ability of the nucleocapsid (N) protein from the SARS-CoV-2 virus to interact with RNA in vitro. Nevertheless, its capacity to interact with deoxyribonucleic acid has yet to be widely described. In the present work, the nucleocapsid protein from the SARS-CoV-2 Delta variant was obtained in E. coli. The recombinant protein was purified, immune identified and incubated with different quantities of the ODN-39M, a CpG ODN with adjuvant properties. As a result, a curve of precipitation was obtained. The analysis by agarose gel electrophoresis of the mixtures revealed that ODN-39M retarded its mobility, whereas the protein increased its migration. When a sample of N+ODN-39M was crosslinked and analyzed by SDS-PAGE, high molecular weight aggregates were detected. Accordingly, upon analysis by Transmission Electron Microscopy, particles of approximately 20 nm were visualized in both samples of N protein, with and without ODN-39M. However, aggregates of particles were abundant in the mixture. Notably, such structures kept the ability of N protein to be recognized by human sera of COVID-19 convalescent donors. We can assert that nucleocapsid protein can bind to deoxyribonucleic acid. Such interaction changed the protein conformation, contributing to the previously reported immunogenicity in mice of N+ODN-39M complexes.
Keywords: Nucleocapsid, SARS-CoV-2, ODN-39M, Particles
INTRODUCTION
In the past 20 years, four lethal coronavirus outbreaks have occurred: SARS (severe acute respiratory syndrome, 2002 and 2003), MERS (Middle East respiratory syndrome, since 2012), and Covid-19 (since 2019). Based on the fact that betacoronaviruses which caused these epidemics are globally distributed in numerous species, the ecologic reality suggests that coronaviruses will emerge again in the future, potentially posing an existential threat. 1
Many current vaccines against SARS-CoV-2 are based on the most variable regions of the virus; therefore, the scope of protection will be limited against future zoonosis events related to coronaviruses.2 To overcome this issue, emerging works are arising to propose potential universal vaccines with a broader scope of protection: the pancorona vaccine.3,4 One of the current strategies is to select conserved regions of the virus as potential antigens for composing vaccine candidates. One of the proteins selected is the Nucleocapsid (N) protein. The N protein is a 45 KDa protein commonly involved in the viral genome's replication, transcription and packaging.5 It is also the most abundant protein in coronaviruses, and its amino acid sequence is usually highly conserved among betacoronaviruses, making this protein a suitable candidate target for a future pancorona vaccine.6 Various works have proven that N protein in different vaccine platforms can protect against SARS-CoV-2 infection in different animal models.7,8 Particularly, our group had previously reported cross-immunity induction in mice, humoral and cellular, when administered as a mixture with the ODN-39M, a CpG ODN with proven adjuvant capacity for other viral proteins. 9,10. It was considered that such immunogenicity was primarily related to the adjuvant nature of the ODN-39M. Nevertheless, a contribution of a possible physical interaction between the N protein and the ODN-39M, which might enhance such cross-immunity, must be considered. It is known that the SARS-CoV-2 N protein, obtained in bacteria, forms biomolecular condensates with RNA in vitro. Such condensates are related to the primary functions of N protein, which are binding to the viral RNA genome and packing them into a long helical nucleocapsid structure or ribonucleoprotein (RNP) complex. Such complexes regulate the viral RNA synthesis in replication/transcription and modulate the infected cell metabolism.11
Based on the above elements, we explored whether the N protein interacts with the ODN-39M in the present work. To do so, the N protein from the SARS-CoV-2 Delta variant was obtained as a recombinant protein in E. coli. The resultant protein was purified (˃90%) and incubated with the ODN-39M. After characterization by different techniques, it was demonstrated that both molecules interact, provoking some changes in the N protein reflected in aggregated particulate structures. Such aggregates show a recognition pattern by COVID-19 convalescent donor sera, similar to that observed for the N protein alone.
MATERIALS AND METHODS
Bacterial strains, ODN-39M and antibodies
Escherichia coli BL21 (DE3): F– ompT gal dcm lonhsdSB(rB- MB-) k(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) was used for expressing the recombinant Nucleocapsid gene.12 For plasmid propagation the plasmid, Escherichia coli strain XL1-blue [F’::Tn10 proAþBþ lacIq D(lacZ)M15/recA1 endA1gyrA96(NaIr) thi hsdR17(rk mþk) supE44 relA1 lac) was used. 13. Antibiotics were added, when necessary, at the following concentrations: Ampicillin (Amp) (100 µg/ml), Kanamycin (Km) (50 µg/ml) and Chloramphenicol (Cl)(34 µg/ml).
ODN-39M: The ODN-39M, a 39 mer, whole phosphodiester backbone ODN (5’-ATC GAC TCT CGA GCG TTC TCG GGG GAC GAT CGT CGG GGG-3'), was synthesized by Sangon Biotech (China).
The anti-N polyclonal antibody developed in rabbits was purchased from Sinobiologicals, China (Cat No. 40588-T62). On the other hand, human sera from COVID-19 convalescent (N=12) and negative (N=10) individuals were collected at Guangdong Eighth People's Hospital. The samples were obtained as part of a study approved by the Institutional Ethics Committee from The Eighth People's Hospital of Dongguan (Guangdong Province, China). Informed written consent was obtained from each participant before enrolment in the study. A more detailed description of the study protocol was previously reported.14
Cloning and expression of the recombinant construct for the Nucleocapsid protein
The nucleocapsid gene of SARS-CoV-2, ID No. YP_009724397.2, which codes for a 1-419 Delta variant, was chemically synthesized and later amplified by polymerase chain reaction (PCR) using the corresponding primers having the restriction sites NcoI/SacI. The amplified band was purified and cloned into pGEM-T Easy Vector (Promega, USA). The restriction analysis tested positive clones, and the Nucleocapsid gene was fully sequenced (Sangon, China). The fragment encoding the N protein was then extracted using NcoI/SacI double digestion and cloned into pET28a plasmid (Sangon, China). The restriction analysis identified positive clones, and the selected clone was conserved under the name pN. The obtained recombinant construct does not contain any tag for further immune detection, purification or characterization.
The E. coli strain BL21 (DE3) was transformed with the pN plasmid by electroporation. N clone was later inoculated, at 0.05 of Optical Density (OD), in ZY medium supplemented with Km (50 µg/ml) and let to grow for 18h at 28°C, with 170 rpm-min (THZ-300, Blue Pard Inst, China). A sample of whole cells was collected to check the expression level.
Purification of the recombinant nucleocapsid protein
ZY medium (1L), supplemented with Kanamycin (50 µg/ml), was inoculated with the bacterial clone expressing the N protein at 0.05 of optical density (OD) and let to grow during 18 h at 28°C, at 170 rpm-min (THZ-300, Blue Pard Inst, China). Later, the cellular biomass was harvested by centrifugation at 5,000 g for 15 min at 40. For cell disruption, 2g of cells were resuspended in 40 mL of the disruption buffer: 0.05M Tris-HCl, 5 mM EDTA, pH 8 (TE buffer). Cells were disrupted at 4°C, with 20 cycles (30 seconds with 30 seconds rest) in a sonicator (FS-200T, Shanghai Sonxi US Inst, China), with 50% power rate and 20.3 kHz frequency. After cell disruption, the sample was centrifuged at 10 000 g for 15 min at 40C, and the soluble fraction was collected and filtered through a 0.45 μm filter for subsequent purification steps.
The ion exchange chromatography was used as the main purification procedure. A volume of 20 mL from the cell disruption soluble fraction was ½ diluted with TE buffer and applied into Q Sepharose fast flow and SP Sepharose fast flow ion exchangers (Cytiva, Sweeden) connected in tandem. Both matrixes were previously equilibrated with TE buffer. The loading volume of the sample was 10%, and the employed flow rate was 3 cm/h. After application, columns were separated, washed independently with TE buffer and the bound proteins to SP Sepharose fast flow matrix eluted with 0.1M NaCl step gradient. The Q Sepharose fast flow matrix was employed to remove most of the protein and DNA contaminants in the sample, while N protein was eluted from the SP Sepharose fast flow matrix with high purity. The N protein fraction was collected at 0.5M NaCl and subsequently filtered by 0.22μm filter.
Gel filtration chromatography was selected as the final polishing purification step. Sephacryl S-200 HR matrix (Cytiva, Sweden) was equilibrated with 0.05 M Tris HCl, 5 mM EDTA, 0.15 M NaCl, pH 8.5, at 15cm/h. Elution from the SP Sepharose fast flow column, at 0.5M NaCl, was applied to the gel filtration chromatography. As a result, only one peak was collected, corresponding to the N protein.
In all chromatographic steps, the detection of the protein signal was followed by measuring the absorbance at 280 nm. SDS-PAGE and Western blotting corroborated the identity of the N protein.
In vitro aggregation procedure of N protein with ODN-39M
As previously described, the purified N protein was subjected to an in vitro aggregation process 15 with few modifications. Briefly, in a 100 µL of reaction, 40 µg of N protein were mixed with different quantities of ODN-39M (2 µg, 4 µg, 8 µg, 16 µg, 40 µg, 60 µg and 80 µg) in 10 mM Tris, 6 mM EDTA (pH 6.9) buffer. The mixtures were incubated for 30 min at 30°C in a water bath, and after, they were stored at 4°C for 4 hours. Finally, each preparation was centrifuged at 14 000 g for 10 min. The soluble fraction was collected and tested. The mixture of 40 µg of N protein and 60 µg of ODN was selected (mass ratio: 0.66) for further studies.
Protein analysis
The protein concentration of samples was determined by BCA assay. In turn, protein samples were subjected to 10-15% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Bands were visualized by staining with Coomassie Brilliant Blue R-250 (Sigma)16
For immune identification, a Western Blot assay was performed. Briefly, protein samples were electrotransferred from an Acrylamide gel to an Immobilon-P membrane (Merck-Millipore, IRL), as described.17 The membrane was blocked with 5% skimmed milk in phosphate buffered saline – Tween solution (PBS-T, PBS and 0.05 % Tween) for 1 h at room temperature (RT), washed three times in PBS-T, and let to react with the anti-N polyclonal Ab generated in rabbit (SinoBiologicals, China) for 1 h at RT. After washing, the membrane was incubated with peroxidase-conjugated goat anti-rabbit IgG (Chemicon, USA) at a 1/300 dilution in PBS-T for 1 h at RT. Afterward, the membrane was rewashed, and the antigen-antibody reaction was visualized by incubation with Aminoethyl Carbazole (AEC) substrate solution (0.2mg/ml 3-amino-9-ethylcarbazole and 0.03 % H2O2 in 50 mM NaAc solution) at room temperature until color development was completed.
The N+ODN-39M mixtures were characterized by Agarose gel electrophoresis (2%), stained with Ethidium Bromide and Coomassie-blue to detect ODN-39M and N protein. The preparation corresponding to 60 µg of ODN-39M (mass ratio N: ODN-39M=0.66) was also treated with 1% formaldehyde (FA) as a crosslinker agent for 5 min at RT. After the treatment, FA-treated samples were quenched by adding Tris to a final concentration 0.1M. SDS-PAGE analyzed samples in non-reducing conditions.
Transmission Electron Microscopy
For microscopy analysis, samples of N protein and N+ODN-39M (mass ratio N: ODN-39M=0.66) were placed on a freshly glow-discharged, 400-mesh copper grid coated with Formvar and Carbon. Uranyl Acetate stain was applied after 2 min of sample absorption and extensive washing with water. After 4 min of staining, grids were wick dried with Whatman no. 1 filter paper and allowed to air dry for 20 min.
Four random fields per sample were observed using a Transmission Electron Microscope (Hitachi system) with an acceleration voltage of 80 Kv and three magnifications: 12 000 x, 25000 x and 60 000 x.
Antigenic characterization using human sera positive to SARS-CoV-2
Ninety-six-well high-binding polystyrene plates (Costar, USA) were coated with 3 μg/mL of N protein (with and without ODN-39M) in sodium carbonate-sodium bicarbonate buffer and incubated overnight at 4°C. Unspecific binding of the antibodies was avoided by blocking with 5% skimmed milk (Oxoid, USA) for 1h at 37°C. After 5 times washing with phosphate-buffered saline with 0.05% Tween 20 (PBST), 100 μL of serum sample diluted (1: 2000) in 2% milk -PBST was added and incubated for 2 hours at 37° C. After washing 5 times with PBST, bound antibodies were detected using a goat anti-human IgG conjugated to horseradish peroxidase [1:20000] (Sigma-Aldrich Co., Germany). After incubation for 1 hour at 37°C and five PBST washes, 100 μL of OPD substrate solution (Sigma-Aldrich Co., Germany) was added to each well and incubated for 10 minutes at room temperature. The reaction was stopped by adding 0.2 N Sulfuric Acid, and the optical density (OD) at 492 nm was measured (FilterMax F3, Molecular Devices, USA). The data is represented as OD measures.
Statistical analysis
The GraphPad Prism version 5.00 statistical software (Graph-Pad Software, San Diego, CA, USA) was used for statistical analyses. The One-way ANOVA test followed by a Tukey's post-test was used as parametric tests for multiple group comparisons. A standard P value consideration was as follows: ns, p>0.05; *, p<0.05; **, p<0.01; ***, p<0.001.
RESULTS
The recombinant nucleocapsid protein of SARS-CoV-2, Delta variant, is successfully expressed in E. coli and can be purified at high purity levels.
The DNA sequence coding for N protein from SARS-CoV-2, Delta variant, was cloned into the PET-28a vector under the plac promoter. E. coli BL21(DE3) was then transformed with the recombinant plasmid and grown at 280C in a ZY auto-induction medium. As shown in Figure 1a, a reinforced band of MW around 45 kDa, corresponding to the theoretical size of the N protein, was visualized, accounting for 5% of the total cellular proteins. In addition, it was adequately immune-identified with polyclonal anti-N Abs, confirming the identity of the recombinant protein obtained (Figure 1b).
After the expression of the recombinant construct, the biomass from the bacterial culture was disrupted. Under established conditions, the recombinant protein was mainly associated with the cell disruption soluble fraction (Figure 1c). This sample was then applied into two ionic exchange chromatographies coupled in tandem (Q and SP Sepharose fast flow). Contaminants were attached to the first anion exchange matrix, whereas the target protein was bound to the cation one. Since the elution from the SP Sepharose was conducted by step gradient, additional contaminants were removed at low molarities of NaCl.
In contrast, the N protein was eluted at 0.5M NaCl with a high purity level. As a final polishing purification step, gel filtration chromatography was introduced using Sephacryl S-200 HR. The chromatography profile and the purified fraction of the last stage of the purification process are shown in Figures 1e and 1f. The protein was obtained with more than 90% of purity.
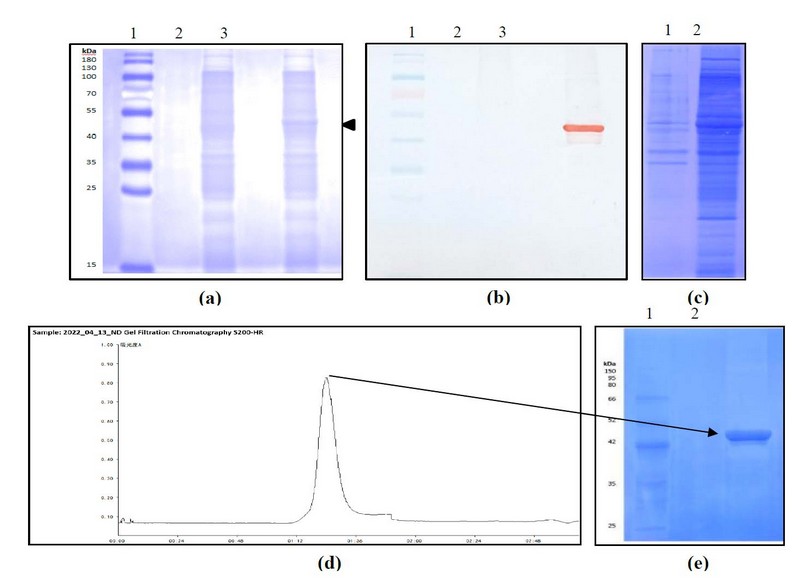
Figure 1. Expression and purification of the recombinant N protein construct. Analysis of expression by (a) SDS-PAGE at 12%, stained with Coomassie Blue, (b) Western blotting using polyclonal anti-N Abs. 1: MW marker, 2: Cell extract of E. coli BL21(DE3) transformed with pET28a, 3: Cell extract of E. coli BL21(DE3) transformed with pN plasmid. (c) Analysis of the disruption process by SDS-PAGE at 12%, stained with coomassie blue 1: Supernatant after disruption, 2: Pellet after disruption. (d) Chromatogram of the gel filtration step. (e) Analysis of the purified protein by SDS-PAGE at 10%, stained with Coomassie Blue. 1: MW marker, 2: Sample eluted from Sephacryl S-200 HR.
The recombinant nucleocapsid protein from the SARS-CoV-2 Delta variant interacts with the ODN-39M
The N protein (40 µg) was subjected to the aggregation process with different quantities of ODN-39M to obtain the aggregation curve (Figure 2a). As a result, maximal precipitation of the mixture was obtained with 4 µg of ODN-39M (mass ration N: ODN-39M=10:1).
To have direct pieces of evidence of protein-ODN-39M interaction, samples of the mixtures were applied into 2% Agarose gel to visualize the localization of each molecule. The gel was stained with Ethidium Bromide to detect ODN-39M and Coomassie-Blue to detect the N protein. As shown in Figure 2b, while higher quantities of ODN-39M are added, the N protein migrated more. At the same time, from the condition of 8 µg of ODN-39M, a smear of ODN-39M appeared (Figure 2c), indicating a retard of the electrophoretic mobility.
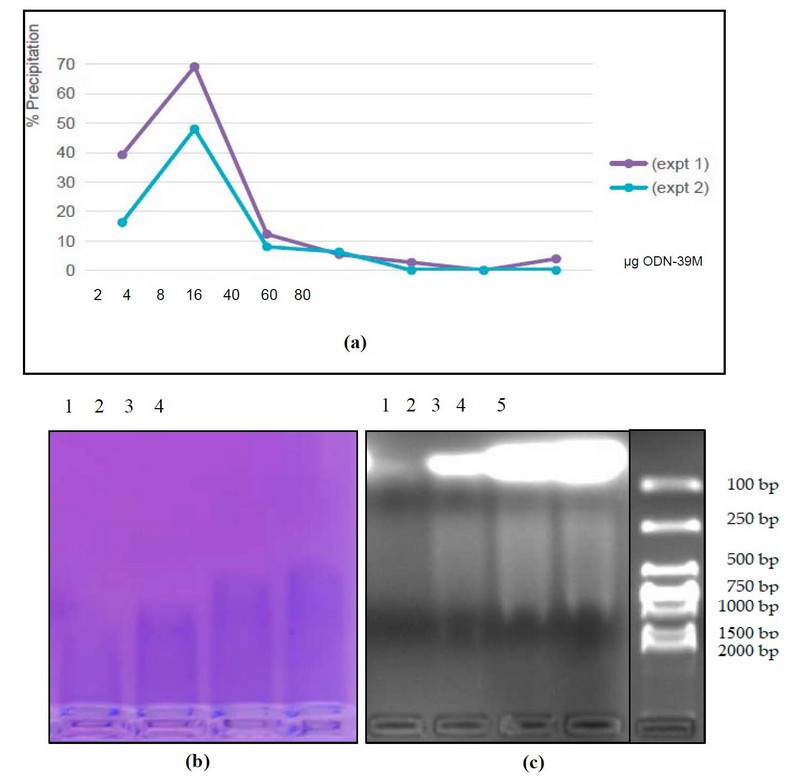
Figure 2. Characterization of the N+ODN-39 M mixture. (a) Precipitation curve of the N protein at different ODN-39M quantities (µg). Two independent experiments are represented. (b) and (c) Analysis of the N+ODN-39M mixture samples by 2% agarose gel electrophoresis stained with (b) Coomassie Blue (c) Ethidium Bromide. 1: N+ODN-39M (2 µg), 2: N+ODN-39M (8 µg), 3: N+ODN-39M (40 µg), 4: N+ODN-39M (60 µg), 5: MW marker (DNA).
The condition corresponding to a mass ratio N: ODN-39M=0.66:1, 60 ugs of ODN-39M, was selected for further characterization of the complexes since precipitation phenomenon did not take place and corresponds to the proportion used in the previous immunological evaluation of a similar preparation, but using a His-tagged N protein.9,10
Protein-ODN complexes (mass ratio N: ODN-39M=0.66:1) were immobilized with 1% Formaldehyde (FA) as a crosslinking agent and further analyzed by SDS-PAGE under non-reducing conditions. The pattern obtained evidenced the presence of high molecular weight N protein aggregates in N+ODN-39M preparation since the sample treated with 1% FA was wholly retained in the gel well (Figures 3a and 3b)
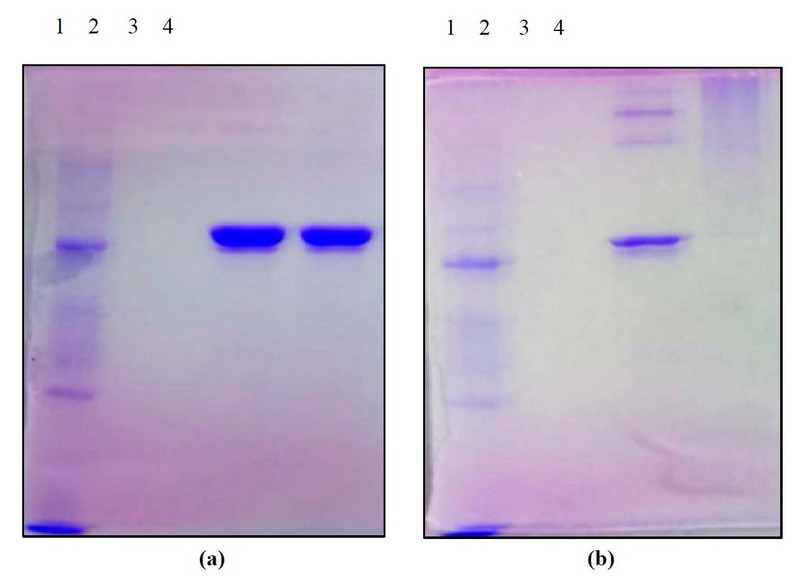
Figure 3. Characterization of samples by crosslinking experiment for the N+ODN-39 M mixture at mass ratio 0.66:1, by SDS-PAGE at 12% (under non-reducing conditions), stained with Coomassie Blue. Samples non-treated (a) and treated with 1% FA as a crosslinking agent (b) 1: MW marker 2: ODN-39M, 3: N protein, 4: N+ODN-39M.
The recombinant N protein forms particles that are more abundant and aggregated upon the addition of ODN-39M
To explore the ability of N protein to form particulate structures, in the presence or not of ODN-39M, both samples, N alone and mixed with ODN-39M (0.66:1 mass ratio), were examined by Transmission Electron Microscopy (TEM). Three pictures, corresponding to three magnification factors of each sample, are shown in Figure 4. Interestingly, particles with a size of approximately 20 nm were visualized in the sample corresponding to the N protein. When the ODN-39 was added, particles of a similar size were more abundant, forming aggregated structures.
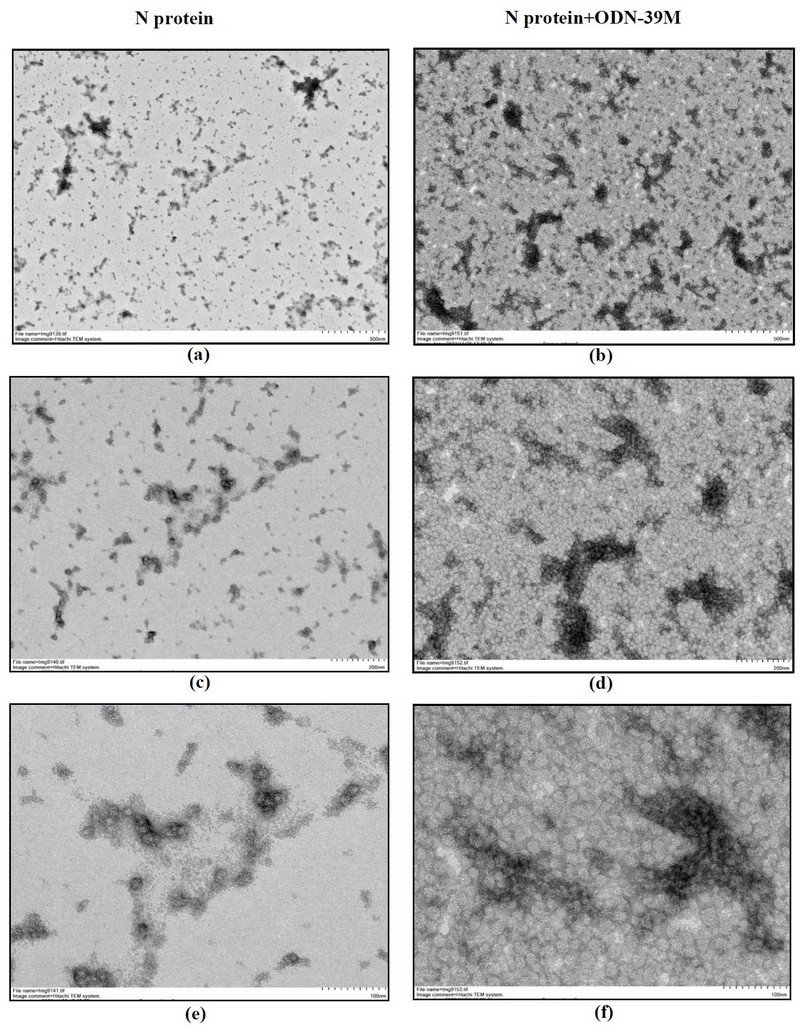
Figure 4. Transmission electron microscopy of N protein (left panel) and N+ODN-39M mixture at mass ratio 0.66:1 (right panel) a, b: magnification 12 000 x (Bar equals 500 nm) c, d: magnification 25 000 x (Bar equals 200 nm) e, f: magnification 60 000 x (Bar equals 100 nm).
The presence of ODN-39 M does not affect the recognition of the nucleocapsid protein by COVID-19 convalescent human sera.
The antigenicity of N protein, without treatment, and after mixture with ODN-39M (mass ratio 0.66:1) was tested using a panel of human sera from COVID-19 convalescent and negative (non-COVID-19 -infected, neither vaccinated) donors, both obtained during an outbreak of SARS-CoV-2 Omicron variant, occurred in 2022, at Guangdong province, China. As shown in Figure 5, both protein preparations were equally recognized by sera from convalescent donors. Results indicate that the addition of ODN-39M did not affect the recognition of N protein by natural anti-SARS-CoV-2 Abs.
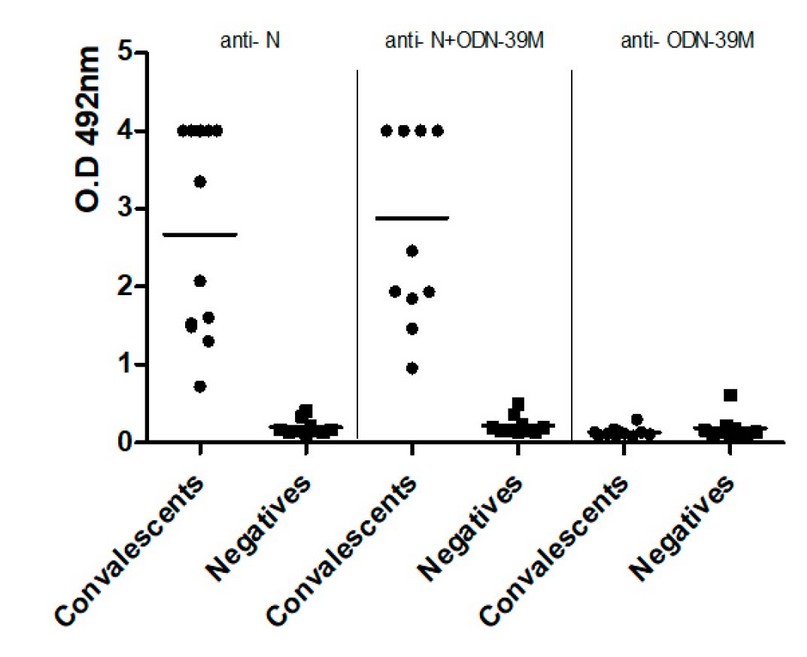
Figure 5. Recognition by human sera of a. N protein (Left panel), N+ODN-39M complexes (middle panel), and ODN-39M (right panel). Data is presented as OD492nm values from each serum. The horizontal bar represents the mean of the group in each case. (●) Convalescents and ( ) negative sera.
DISCUSSION
Given that one function of the SARS-CoV-2 nucleocapsid protein is to package the viral genome into a ribonucleic nucleoparticle, and it can phase separates with viral and non-viral RNA sequences, we hypothesized that N protein could also interact with the ODN-39M to form aggregated structures.18
The present study obtained a recombinant His-tag-free N protein, considering vaccine formulation requirements. The Delta variant was selected since it was the significant circulating strain when these studies started. Despite Delta wave being overtaken by subsequent variants, this variant was considered one of the most SARS-CoV-2 aggressive strains, and its immunogenicity has not been deeply explored in the context of vaccine candidates.
The recombinant construct pN was successfully expressed in E. coli, and the recombinant protein was obtained in the soluble fraction after cellular disruption, pointing to its proper conformation. The developed purification process rendered a protein preparation with more than 90% purity, suitable for the subsequent aggregation reaction with the ODN-39M.
The aggregation (precipitation) curve, obtained upon the mixture of the N protein with different quantities of ODN-39M, reveals an interaction between both molecules. Similar curves were obtained by Jack et al., 2021 when combined with the N protein obtained in mammalian cells with different variants of RNA.11 The maximal point of precipitation in the present study, at mass ratio N: ODN-39M 10:1 (4 µg of ODN-39M), reveals that N protein is also able to phase separate upon interaction with a limited amount of a DNA fragment (ODN-39M) in a similar way to that reported for different RNAs. On the other hand, analyzing the N: ODN-39M samples, we found that the migration of the N protein in a 2% agarose gel depends on the quantity of ODN-39M, and at the same time, a retarded mobility of ODN-39M was observed. These signals reveal that the N protein interacts with the ODN-39M, forming complexes, constituting a second evidence of interaction between those molecules. The same migration delay for the nucleic acid was visualized when a recombinant N protein, also obtained in E. coli, was mixed with three RNA fragments (400, 600, and 800 nt) from the 5' end of the SARS-CoV-2 genome. All three RNAs shifted to a larger species in the presence of the N protein. 19 Similarly, Qiaozhen et al., 2021 evidenced an interaction between the N recombinant protein and a 7-mer single-stranded RNA or dsRNA. 18
Even though the N protein from SARS-CoV-2 has been focused as an attractive target for vaccine development, its capacity to form particles has not been deeply studied. In the present work, N protein, as well as the sample corresponding to complexes at mass ratio N: ODN-39M of 0.66:1, were characterized by crosslinking experiment and TEM, since this was the condition previously evaluated in mice with successful results, but using the His-tagged N protein. 9,10 In this condition, the ODN is not limiting; according to the electrophoretic pattern, we estimate that only around 10 - 20% of the total amount of ODN-39M in the mixture was effectively bound to the N protein in this condition.
According to the SDS-PAGE electrophoretic pattern of the samples treated with 1% FA, we detected that around 50% of the N protein formed high molecular weight aggregates, which follows the reported ability to self-associate into tetramers and higher oligomers of the N protein of other coronaviruses. 20,21 The level of self-association of the N protein from SARS-CoV-2 in the present work was also confirmed by TEM since spherical particles of approximately 20 nm were visualized. In turn, when ODN-39M was added, high molecular weight structures were visualized by SDS-PAGE. Accordingly, abundant 20 nm spherical particles were also visualized by TEM, forming aggregated structures. This constitutes a third evidence supporting an interaction between the recombinant N protein from SARS-CoV-2 and the ODN-39M.
Of note, the morphology of particles visualized in our study differs from that described by Carlson et al., 2022. The authors incubated the N protein with 600 nt RNA and visualized cylindrical 15 nm particles, similar to the vRNP structures observed within coronavirus virions.19 Such morphological differences might be explained by 1) the length of each nucleic acid: 600 nt vs. 39 nt and 2) the nature of the nucleic acid in each complex: RNA vs. DNA. Despite the morphological differences, the diameter of the particles is very close, 15 nm for RNP vs. 20 nm for the particles visualized in the present work.
Since recent pieces of evidence support that Abs generated against N protein may contribute to the control of the virus infection, 22,23 we determined whether the addition of ODN-39M would affect the antigenicity of the N protein. The similar level of recognition found for both samples, N protein with and without ODN-39M, revealed that the addition of ODN-39M did not affect the ability of N protein to be recognized by human sera of COVID-19 convalescent donors.
Taken together, we hypothesize that a small portion of the ODN-39M bound to N protein (condition 0.66:1 protein/ODN mass ratio) provokes a conformational change in it, which is reflected in the increase of the particle quantity as well as the formation of high molecular weight structures, visualized as aggregates of particles by TEM.
CONCLUSIONS
Results demonstrate that Nucleocapsid protein from SARS-CoV-2 (Delta variant) can be obtained as a recombinant protein in E.coli with a high purity level and a proper conformation. It can interact with the ODN-39M, forming aggregate structures preserving antigenicity. These findings and the importance of N protein as an antigen are relevant in designing novel and broader Coronavirus vaccine candidates. Further studies are ongoing to evaluate the immunogenicity of the aggregate structures obtained in the present work.
Author Contributions: Conceptualization, LH, YL, and YP; methodology, YL, A.M., ES, VH and LH; validation, LH and YP; formal analysis, LH, YL, YP, A.M., VH and RS; investigation, YL, A.M., ES, RC, PA and CT; resources, LH and KY; data curation, YL, A.M., LH, ES and RS; writing—original draft preparation, LH and YL; writing—review and editing, LH, YL, RS and YP; supervision, LH and YP; project administration, LH, YP and KY; funding acquisition, LH, YP and KY All authors have read and agreed to the published version of the manuscript
Funding: This work was supported by MOST "National key R&D program of China (2021YFE0192200)", "PNCT CITMA, Cuba", "Hunan Provincial Base for Scientific and Technological Innovation Cooperation (2019CB1012)", "The Science and Technology Innovation Program of Hunan Province, (2020RC5035)", "Hunan Provincial Innovative Construction Program (2020WK2031)" The APC was funded by MOST "National key R&D program of China (2021YFE0192200)". The APC was funded by MOST "National Key R&D Program of China (2021YFE0192200)."
Data Availability Statement: This section details where data supporting reported results can be found, including links to publicly archived datasets analyzed or generated during the study. Please refer to the suggested Data Availability Statements in the "Bionatura Research Data Policies" section at https://www.revistabionatura.com/policies.html. You might exclude this statement if the study did not report any data.
Acknowledgments: We thank Dr. Jiang from Guangdong Eighth People's Hospital, Guangdong Province, China, for providing the samples from COVID-19 convalescent donors.
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the study's design, in the collection, analysis, or interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
REFERENCES
1. Morens, DM; Taubenberger, J.K.; Fauci A.S. A centenary tale of two pandemics: the 1918 influenza pandemic and COVID-19, part II. Am J Public Health 2021, 111: 1267-72.
2. Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O'Connell, A.-M.; et al. Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N Engl J Med 2022, 386: 1532–1546. doi:10.1056/NEJMoa2119451
3. Rubin, R. The Search for a Single Vaccine Against Coronaviruses Yet to Come. JAMA 2021, 326(2):118-120. https://doi:10.1001/jama.2021.9477
4. Morens, D.M; Taubenberger, J.K.; Fauci, A.S. Universal Coronavirus Vaccines — An Urgent Need. N Engl J Med 2022, 386: 297–299. https://doi.org/10.1056/NEJMp2118468
5. Wu, W.; Cheng, Y.; Zhou, H.; Sun, C.; Zhang, S. The SARS‑CoV‑2 nucleocapsid protein: its role in the viral life cycle, structure and functions, and use as a potential target in the development of vaccines and diagnostic. Virology Journal 2023, 20:6. https://doi.org/10.1186/s12985-023-01968-6
6. Dutta, N.K; Mazumdar, K.; Gordy, J.T. The Nucleocapsid Protein of SARS–CoV-2: a Target for Vaccine Development. J Virol 2020, 94: e00647-20. https://doi.org/10.1128/JVI.00647-20
7. Matchett, W.E.; Joag, V.; Stolley, J.M.; Shepherd, F.K.; Quarnstrom, C.F.; Mickelson, C.K.; Wijeyesinghe, S.; Soerens, A.G.; Becker, S.; Thiede, J.M.; Weyu, E.; O'Flanagan, S.D.; Walter, J.A.; Vu, MN; Menachery, V.D.; Bold, TD; Vezys, V.; Jenkins, M.K.; Langlois, R.A.; Masopust, D. Cutting Edge: Nucleocapsid Vaccine Elicits Spike-Independent SARS-CoV-2 Protective Immunity. J Immunol 2021, 207: 376–379. https://doi.org/10.4049/jimmunol.210042
8. Dangi, T.; Class, J.; Palacio, N.; Richner, J.M.; Penaloza MacMaster, P. Combining spike- and nucleocapsid-based vaccines improves distal control of SARS-CoV-2. Cell Reports 2021, 36: 109664. https://doi.org/10.1016/j.celrep.2021.109664
9. Lobaina, Y,; Chen, R.; Suzarte, E.; Ai, P.; Huerta, V.; Musacchio, A.; Silva, R.; Tan, C.; Martin, A.; Lazo, L.; Guillén, G.; Yang, K.; Perera, Y.; Hermida, L. The Nucleocapsid Protein Of SARS-CoV-2, Combined With ODN-39M, Is A Potential Component For An Intranasal Bivalent Pancorona Vaccine. (preprint) Immunology 2022. https://doi.org/10.1101/2022.06.02.494502
10. Lobaina, Y, Chen, R.; Suzarte, E.; Ai, P.; Huerta, V.; Tan, C.; Alvarez-Lajonchere, L.; Liling, Y.; Musacchio, A.; Silva, R.; Guillén, G.; Yang, K.; Perera, Y.; Hermida, L. Broad humoral immunity generated in mice by a formulation composed by two antigens from delta variant of SARS-CoV-2. Arch of Virol 2023.(accepted).
11. Jack, A.; Ferro, L.S.; Trnka, M.J.; Wehri, E.; Nadgir. A.; Nguyenla, X.; Fox, D. ; Costa, K.; Stanley, S.; Schaletzky, J.; Yildiz A. SARS-CoV-2 nucleocapsid protein forms condensates with viral genomic RNA. PLoS Biol 2021, 19(10): e3001425. https://doi.org/10.1371/journal.pbio.3001425
12. Wood, W.B. Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol 2011, 16:118–133.
13. Bullock, W.O.; Fernandez, J.M.; Short, J.M.S. XL1-Blue: a high-efficiency plasmid transforming recA Escherichia coli strain with Beta-Galactosidase selection. Biotechniques 1987, 5:376–378.
14. Lobaina, Y.; Chen, R.; Ai, P.; Yang, L.; Alvarez-Lajonchere, L.; Suzarte, E.; Tan, C.; Silva, R.; Jiang, Z.; Yang, K.; Perera, Y.; Hermida, L. Cross-Reactive Profile Against Two Conserved Coronavirus Antigens in Sera from SARS-CoV-2 Hybrid and Vaccinated Immune Donors. Viral Immunol 2023, 36(3): 222–228. https://doi.org/10.1089/vim.2022.0186
15. Gil, L.; Marcos, E.; Izquierdo, A.; Lazo, L.; Valdés, I.; Ambala, P.; Ochola, L.; Hitler, R.; Suzarte, E.; Álvarez, M.; Kimiti, P.; Ndung’u, J.; Kariuki, T.; Guzmán, M.G.; Guillén, G.; Hermida, L. The protein DIIIC‐2, aggregated with a specific oligodeoxynucleotide and adjuvanted in alum, protects mice and monkeys against DENV‐2. Immunol Cell Biol 2015, 93: 57–66. https://doi.org/10.1038/icb.2014.63
16. Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227: 680–685.
17. Towbin, H.; Staehelin, T.; Golden, J. Electrophoretic transfer of protein from polyacrylamide gel to nitrocellulose sheets procedure and some applications. Proc Natl Acad Sci 1979, 76: 4350–4354.
18. Ye, Q.; Lu, S.; Corbett, K.D. Structural Basis for SARS-CoV-2 Nucleocapsid Protein Recognition by Single-Domain Antibodies. Front Immunol 2021, 12: 719037.doi: 10.3389/fimmu.2021.719037
19. Carlson, C.R.; Adly A.M.; Bi, M.; Cheng, Y.; Morgan, D.O. Reconstitution of the SARS-CoV-2 ribonucleosome provides insights into genomic RNA packaging and regulation by phosphorylation. Preprint 2022. https://doi.org/10.1101/2022.05.23.493138
20. Chen, C.Y.; Chang, C.K.; Chang, Y.W.; Sue, S.C.; Bai, H.I.; Riang, L.; et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol 2007, 368(4):1075–86. https://doi.org/10.1016/j.jmb.2007.02.069
21. Chang, C.K.; Chen, C.M.; Chiang, M.H.; Hsu, Y.L.; Huang, T.H.; Transient oligomerization of the SARS-CoV N protein—implication for virus ribonucleoprotein packaging. PLoS ONE 2013, 8(5):e65045. https://doi.org/10.1371/journal.pone.0065045
22. Dangi, T.; Sanchez, S.; Park, M.; Class, J.; Richner, J.M.; Penaloza-MacMaster, P. Nucleocapsid-specific humoral responses improve the control of SARS-CoV-2. Preprint 2022. https://doi.org/10.1101/2022.03.09.483635
23. Herman, J.D.; Wuang, C.; Burke, J.S.; Zur, Y.; Compere, H.; Kang, J.; Macvicar, R.; Shin, S.; Frank, I.; Siegel, D.; Tebas, P.; Choi, G.H.; Shaw, P.A.; Yoon, H.; Liise-anne Pirofski, L-A.; Juelg, B.; Bar, K.J.; Lauffenburger, D.; Alter, G. A role for Nucleocapsid-specific antibody function in Covid-19 Convalescent plasma therapy. Preprint 2022. https://doi.org/10.1101/2022.02.19.22271230
Received: 28 May 2023/ Accepted: 15 July 2023 / Published:15 September 2023
Citation: Lobaina Y, Musacchio A, Ai P, Chen R, Suzarte E, Tan C, Silva R, Yang K, PereraY, Hermida L Cloning, expression, purification and characterization of the recombinant nucleocapsid protein from SARS-CoV-2 and its combination with a CpG ODN-39M. Revis Bionatura 2023;8 (3) 21. http://dx.doi.org/10.21931/RB/2023.08.03.21
