2021.06.03.17
Files > Volume 6 > Vol 6 No 3 2021
INVESTIGATION / RESEARCH
Synthesis, in silico studies and antibacterial assessment of α-amino phosphonates derivatives
Bouchra El Khalfi1, Boutaina Addoum1, Suhayla Harrati1, Abdelhakim Elmakssoudi2 and Abdelaziz Soukri1*
Available from: http://dx.doi.org/10.21931/RB/2021.06.03.17
ABSTRACT
The widespread of multi-resistant strains due to the lack of specific treatment and the propagation of infectious diseases requires all resources to remedy this scourge. This study is therefore aimed to assess the antibacterial activity of four synthetic α-Aminophosphonate 4(a-d). Methods: Firstly, α-Aminophosphonate has been synthesized and characterized, then molecular docking of these compounds 4(a-d) into the active binding site of Escherichia coli MurB enzyme (PDB Id: 1MBT) was performed to gain a comprehensive understanding of their biological activity. These compounds have been subjected to in vitro antibacterial screening against three multi-resistant strains E. coli, S. aureus, and L. monocytogenes. These compounds showed crucial antibacterial behavior against all studied strains. Thus, their docking estimation supported the in vitro results and showed that the 4c derivative has considerable binding energy towards the active site of Escherichia coli MurB. These findings provide critical information for the exploration of α-amino phosphonates as novel antibacterial agents.
Keywords: α-Aminophosphonate, Docking, Antibacterial.
INTRODUCTION
The emergence and widespread bacterial resistance to antibiotics dramatically sapped our ability1 to remedy this infection and precipitate an alarming public health dilemma2. Nowadays, the new obsession of researchers is to innovate new therapeutic approaches able to overcome these microbial diseases3. Several studies have been focused on the synthesis of eco-friendly substances4. Among these molecules, α-amino phosphonates take a leading position5, especially in biological and environmental applications6–9.
These compounds attracted the renewed interest of biologists due to their wide-ranging activities; they are recognized as an overwhelming inhibitor of enzymes like serine10, UDP- galactopyranose mutase 11, and the viral enzyme human immunodeficiency virus protease. Likewise, they limited metastatic progression, activated different apoptosis pathways, and induced anti-tumoral cytotoxicity.
In this regard, the current study pertains to the synthesis and characterization of some α-amino phosphonates derivatives 4(a-c), then we evaluated in silico their antimicrobial potential 12 based on a computer-aided simulation 13. Then, we tested their antibacterial behavior on resistant strains and their synergistic effect with some standard antibiotics.
All this investigation is essential to understand better the mechanism of action of these molecules, which could offer an exceptional framework that may lead us to the discovery of new potent antibiotics.
METHODS
The general protocol of α-amino phosphonates synthesis
The multicomponent synthesis of the 4 derivatives of α-amino phosphonates was carried out through the well-established literature protocol14,15. We grafted an aromatic aldehyde (1a), aniline (2), and diethyl phosphite (3) in the presence of Na2CaP2O7 as a catalyst16. A plausible reactional mechanism of the one-pot domino reaction is illustrated in Scheme S1of ESI.
Characterization of th α-aminophosphonates 4(a-d)
Characterization of the Diethyl(phenyl)-N-(phenyl) aminomethylphosphonate 4a
1H-NMR (CDCl3) δ: 1.2 (3H, JHH= 7.2 Hz, t, OCH2CH3); 1.4 (3H, JHH= 7.2 Hz, t, OCH2CH3); 3.73-4.3 (4H, m, OCH2CH3); 4.9 (1H, JHP= 24.6 Hz, d, CHP), 5 (1H, s, NH); 6.6-7.8 (10H, m, HAr).13C-NMR (CDCl3) δ: 16.46 (d, 3JCP= 6.03 Hz, OCH2CH3); 16.7 (d, 3JCP= 6.03 Hz, OCH2CH3); 56.35 (d, 1JCP = 149 Hz, CHP); 63.5 (d, 2JCP = 6.79Hz, OCH2CH3); 63.53 (d, 2JCP = 6.79 Hz, OCH2CH3); 114.13 (s); 118.64 (s); 128.1 (s); 128.85 (s); 129.43 (s); 136.2 (s); 146.6 (s). IR (KBr) υ 3304(NH), 2985(CH), 1605 (C=C), 1514 (C=C), 1240 (P=O),1020 (P-O) cm-1.
Diethyl(4-methoxyphenyl)-N-(phenyl) aminomethylphosphonate 4b
1H-NMR (CDCl3) δ : 1.25 (3H, JHH= 7.2 Hz, t, OCH2CH3); 1.4 (3H, JHH= 7.2 Hz, t, OCH2CH3); 2.45 (3H, s, C6H4CH3);3.79-4.31 (4H, m, OCH2CH3); 4.87 (1H, JHP= 24.6 Hz, d, CHP), 5 (1H, s, NH); 6.71-7.5 (9H, m, HAr).13C-NMR (CDCl3) δ: 16.5 (d, 3JCP= 5.8 Hz, OCH2CH3); 16.71 (d, 3JCP= 5.8 Hz, OCH2CH3); 21.4 (s, C6H4CH3); 56 (d, 1JCP = 150 Hz, CHP); 63.48 (d, 2JCP = 6.94 Hz, OCH2CH3); 114.14 (s); 118.57 (s); 128 (s); 129.4 (s); 129.55 (s); 129.58 (s); 133 (s); 137.8 (s);146.8 (s). IR (KBr) υ 3325 (NH), 2980 (CH), 1604 (C=C), 1498 (C=C), 1234 (P=O), 1016 (P-O) cm-1
Characterization of the Diethyl(2-hydroxyphenyl)-N phenyl) Aminomethylphosphonate 4c
1H-NMR (CDCl3) δ : 1.25 (3H, JHH= 7.2 Hz, t, OCH2CH3); 1.39 (3H, JHH= 7.2 Hz, t, OCH2CH3); 3.86 (3H, s, C6H4OCH3);3.8-4.26 (4H, m, OCH2CH3); 4.85 (1H, JHP= 23.1 Hz, d, CHP), 4,95 (1H, s, NH); 6.7-7.53 (9H, m, HAr).13C-NMR (CDCl3) δ: 16.53 (d, 3JCP= 5.7 Hz, OCH2CH3); 16.72 (d, 3JCP= 5.7 Hz, OCH2CH3); 55.44 (s, C6H4OCH3); 55.62 (d, 1JCP = 151,2 Hz, CHP); 63.43 (d, 2JCP = 6.9 Hz, OCH2CH3); 63.47 (d, 2JCP= 6.9 Hz, OCH2CH3);114.15 (s); 114.32 (s); 118.57 (s); 128 (s); 129.2 (s); 129.33 (s); 146.67 (s); 159.56 (s). IR (KBr) υ 3300 (NH), 2983 (CH), 1600 (C=C), 1510 (C=C), 1232 (P=O), 1020 (P-O) cm-1.
Characterization of the Diethyl(4-methylphényl)-N(phenyl)aminomethylphosphonate 4d
1H-NMR (CDCl3) δ : 1.25 (3H, JHH= 7.2 Hz, t, OCH2CH3); 1.39 (3H, JHH= 7.2 Hz, t, OCH2CH3); 3.86 (3H, s, C6H4OCH3);3.8-4.26 (4H, m, OCH2CH3); 4.85 (1H, JHP= 23.1 Hz, d, CHP), 4,95 (1H, s, NH); 6.7-7.53 (9H, m, HAr).13C-NMR (CDCl3) δ: 16.53 (d, 3JCP= 5.7 Hz, OCH2CH3); 16.72 (d, 3JCP= 5.7 Hz, OCH2CH3); 55.44 (s, C6H4OCH3); 55.62 (d, 1JCP = 151,2 Hz, CHP); 63.43 (d, 2JCP = 6.9 Hz, OCH2CH3); 63.47 (d, 2JCP= 6.9 Hz, OCH2CH3);114.15 (s); 114.32 (s); 118.57 (s); 128 (s); 129.2 (s); 129.33 (s); 146.67 (s); 159.56 (s). IR (KBr) υ 3300 (NH), 2983 (CH), 1600 (C=C), 1510 (C=C), 1232 (P=O), 1020 (P-O) cm-
Biological experimental data
Computational studies
Determination ADMET parameters
The ADMET parameters (absorption, distribution, metabolism, excretion, and toxicity of the synthesized compounds 4(a-d) were calculated using the freely accessible web server Swiss ADME (http://swissadme.ch/index.php#undefined)17
Protein-Ligand Docking calculations
After preparing protein/ligands structures, we stimulated the docking interaction using the software AutoDock 1.5.6 (MGL tools- 1.5.6)18. Polar hydrogen atoms and Kollman united charges were added to all target proteins, and the resulting file has been saved in pdbqt extension. For the docking calculation, a grid box of 60×60×60 Å in x, y, z directions were created to cover the active site of the target. The default grid points spacing was fixed to 0.375 Å and centred at x = 42.527 y = -46.679, z = 65.559. We used in this work the Lamarckian Genetic Algorithm (LGA) for flexible docking calculations. The LGA parameters including size, energy screening, mutation rate, and crossover rate. After the calculation procedure, we selected the best conformations of the complex from the 10 calculated based on their binding energy scores. Then the complex interaction map was displayed using Discovery Studio Visualizer17,19.
Microbiology
Bacterial strains
Three bacterial strains E. coli, S. aureus, and L. monocytogenes have been used in this study. The media used for antibacterial screening was the LB and the Brain Heart Infusion (BHI).
Preparation of stock solutions
To prepare a final concentration of 5 μg / ml, we solubilized the selected compounds in DMSO (16.66 μg / ml). Then the stock solutions were stored in sterile containers in obscurity.
Antibiotics
Stocks of antibiotics: ampicillin (100 mg / ml), chloramphenicol (34 mg / ml) and rifampicin (50 mg / ml) were prepared, filtered and stored at -20 ° C until microbiological assays. The chloramphenicol is prepared in ethanol and rifampicin was formulated in methanol.
Antibacterial sensitivity test
The compounds 4(a-d) were tested for their antibacterial activity against E.Coli; S.aureus, and L.monocytogene by using the agar well diffusion method20. To explore the antibacterial effect of these compounds, we subculture the different strains by streaking the supercooled media (LB and BHI) with bacterial inoculum adjusted to 106 CFU (bacteria/ml) then poured into Petri dishes. After 15 minutes, wells of 6mm were made on each plate by using a sterile cone. Under aseptic conditions, we injected separately 50 μl of the tested molecule; positive control (antibiotic), and negative control (DMSO) in each well. The Petri dishes were incubated at 30 - 37 ° C for 24h. All tests were performed in triplicate.
Test of synergistic effect
We used the diffusion assay described previously to study the synergistic effect between standard antibiotics and the target compounds. Firstly, wells were produced in each plate and filled with 50 μl of the tested samples containing the mixture of molecules and antibiotics. The Petri dishes are maintained at 30 ° - 37 ° C for 24 hours. The use of antibiotics singly serves as a positive control and DMSO is considered a negative control. All tests were obtained in triplicate.
RESULTS
ADMET studies
The ADMET data provides information on the lipophilicity of the ligands, which is predicted by the MIlog P parameter, the hydrogen bonding potential of the compounds, and their molecular flexibility21. The different drug-likeness proprieties for all selected compounds were in harmony with Lipinski's rule of five 12 (see table 1).
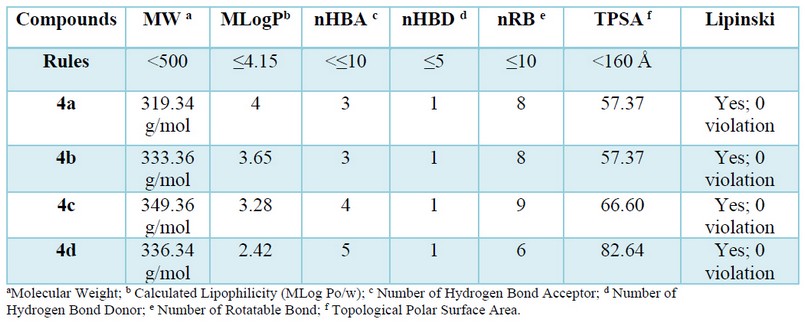
Table 1. Determination of pharmacokinetic parameters for good oral bioavailability of synthesized compounds 4(a-d).
Binding energy evaluation
Modeling studies are essential to understand the mechanism of action of these designed compounds. Our Docking investigation has been confirmed that the highest energies of binding (ΔG) observed are -6.44 and -6.45 K. cal/mol for the 4c and the 4a (see table 2), which can reflect the good affinity between the protein targeted and ligand in contrast to the ligand of reference (ciprofloxacin).
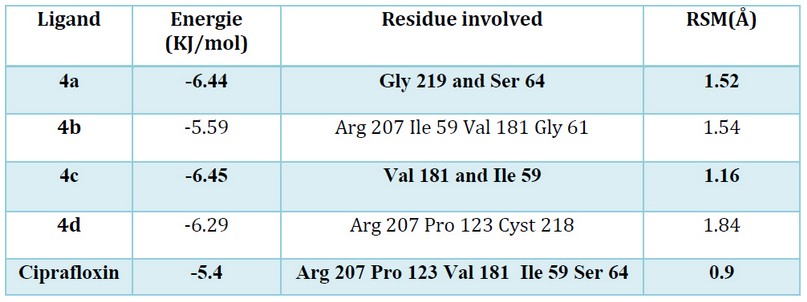
Table 2. The binding score of the tested compounds docked into the active site of 1MBT.
All the docked molecules were subjected to 2D protein-ligand interaction analysis and the obtained conformations compared to the Ciprafloxin have been reported in Figures 3. The selected compounds complex to the target enzyme (MurB) represented at least two H-bonds. Accordingly, the docking studies of α-amino phosphonates gathered previously in table 2 revealed their free energy of binding to E. coli UDP-N-acetylenolpyruvoylglucosamine reductase (MurB) was higher for all compounds than the score obtained for standard antibiotic (Ciprofloxacin). Hence, we can conclude that E. coli MurB is the putative target responsible for the antibacterial activity of these compounds22. Their interaction with the target was stabilized by forming hydrogen bonds, hydrophobic interactions, Van der walls Pi-sulfur and Pi-alkyl interactions (Figure 3).
Additionally, the binding mode of the most active compound 4c (binding energy: -6.45 kcal/mol) showed two hydrogen bonds formed between the oxygen atom and the amino acid Ile 59. We also detected the formation of close interaction with Val 181 via a hydrogen bond. Moreover, Pi-alkyl and Pi-sulfur interactions are formed with the residues Leu 80Lys 292 and Met 132. These amino acids reside at the enzyme's active site and are crucial in the biosynthesis of peptidoglycan; any modifications in these amino acids could down-regulate the enzyme activity, which induces bacterial cell death23.
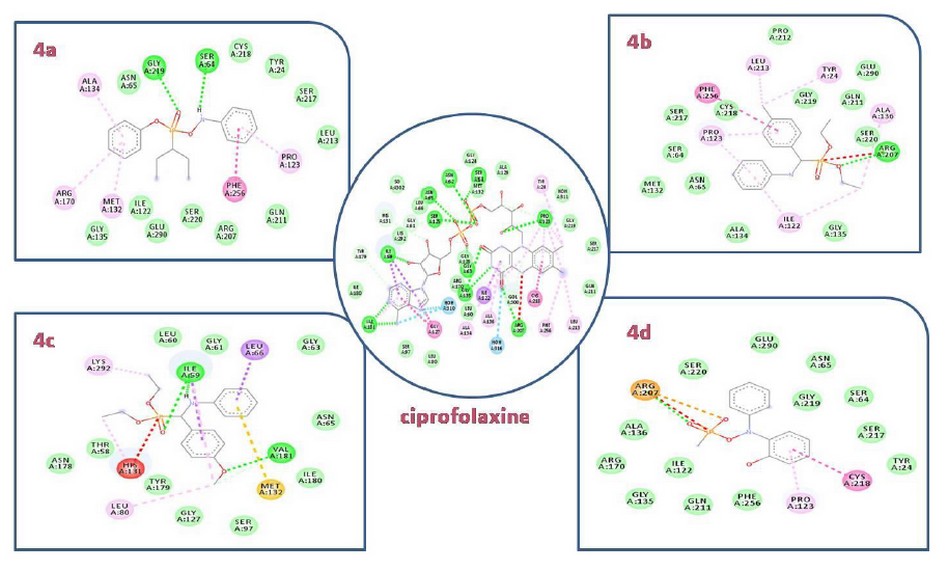
Figure 3. 2D docking diagrams of compounds 4a, 4b, 4c and,4d at the active site of MurB
In vitro studies
The antibacterial effect of the four synthesized compounds against the three strains was investigated by using the diffusion method, and the results are reported in Table 3. The obtained data showed clearly that the 4c compound possesses significant activity against the whole strains compared to the other derivatives. We also observed that the 4a and 4b were more actives against E. coli and S. aureus, while the 4d compound showed a substantial activity only against L. monocytogenes. On the other side, no inhibition zone was detected with vehicle control (DMSO).
Synergistic Test
The effect of the combination between the 4 synthesized products and antibiotics viz ampicillin, chloramphenicol, and rifampicin on the viability of the strain are summarized in figures 1 and 2.
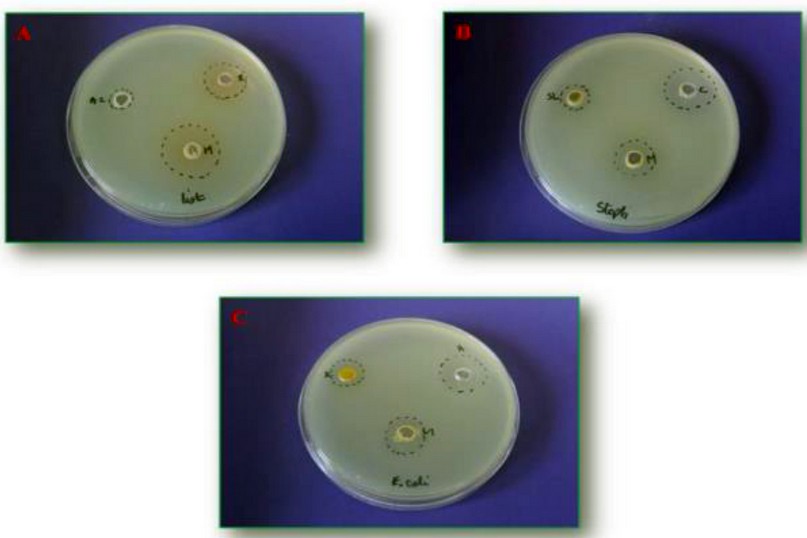
Figure 1. Synergistic effect between α-amino phosphonates and commercial antibiotics. A: a mixture between rifampicin and compound 4b, B: chloramphenicol supplemented to 4c, and C: ampicillin added to the compound 4c. *M: molecule/antibiotic, List: L. monocytogenes, Staph: S. aureus
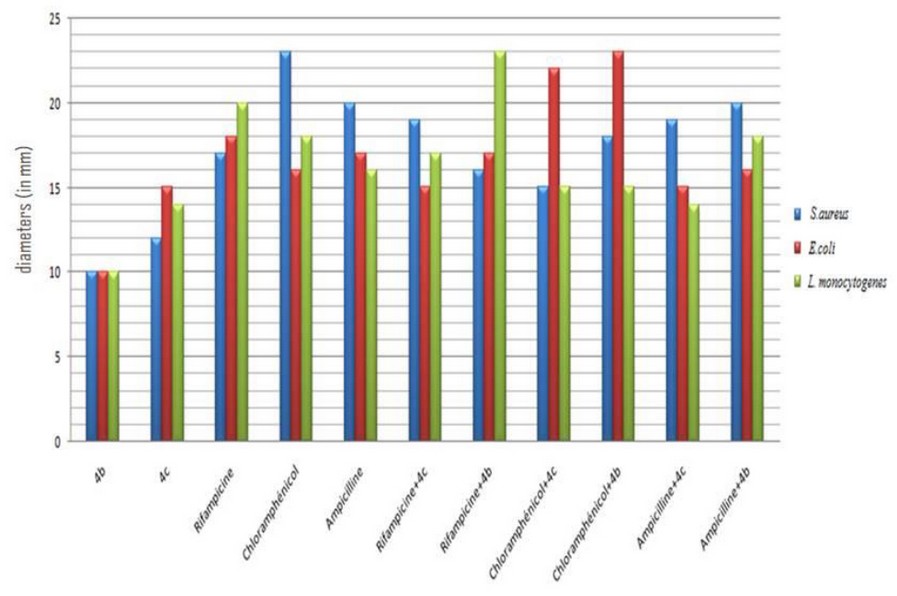
Figure 2. The plot of a synergistic effect of α-amino phosphonates/antibiotic on the growth inhibition of studied strains
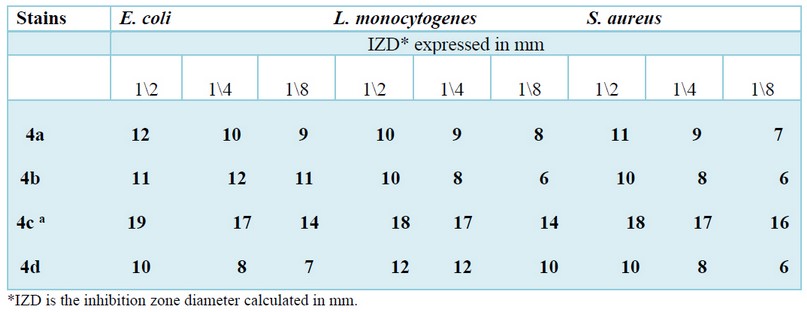
Table 3. Antibacterial activity of the synthesized compounds 4(a-d).
By comparing the above results, it's exciting to underline that the synergistic effect of the synthesized products varies as a function of the studied strain and the antibiotic used. The potent inhibition was detected against S. aureus and L. monocytogenes after rifampicin to the 4 b and 4c compounds. Nevertheless, when we combine ampicillin/4b, chloramphenicol/4b, and chloramphenicol/4c we obtain a sizable activity only against E. coli (G- bacteria).
DISCUSSIONS
The in silico data have been supported our study in vitro performed previously against Multi-resistant strains. According to our docking finding, all the selected compounds had H-bond interactions with the MurB enzyme indicating that the binding affinity increased significatively with the number of the H-bond, which reflects the stability of the complex ligand-protein.
We also observed that the studied compounds fulfill Lipinski's rule requirements and represent a high binding score with an RSM (root square mean) value < 2Å, which confirms their antibacterial inhibiting activity and the reliability of our virtual screening24. Based on molecular docking analysis, we can conclude that the putative mechanism of antibacterial activity observed in vitro is probably the inhibition of the E. coli MurB enzyme. This enzyme catalyzed the second step in forming muramyl sugar 22, which is essential for peptidoglycan biosynthesis25,26.
On the other side, the sensitivity of the studied strains against these molecules provides their antibacterial potency estimated using an inhibition zone formed around the well. If we should classify these molecules as a function of their increasing antibacterial potential order: the 4c compound takes the place position due to their great effect against the three multi-resistant strains viz S. aureus, E. coli et L. monocytogenes(4c>4b> 4a >4d). At the same time, the three other compounds possessed a variable antibacterial activity as a function of the strain used. For example, the 4a and 4b derivatives possess antibacterial activity against gram-negative strains (E. coli); Nevertheless, the 4d derivative is more efficient against gram-positive strains(L. monocytogenes and S.aureus). It's gratifying to elucidate that this antibacterial sensitivity presumably related to the structure of the tested compounds, as described the substitution of bioactive motive such methyl previously; methoxy, or metoxybenzaldhyde in the phenyl ring enhance this activity27; as results when the aromatic aldehyde is replaced with hydroxyl scaffold, we notice a slight and inconspicuous antibacterial activity28,29.
Our results are in harmony with some previous studies. It's gratifying to cite the research work published in 201130, and they showed the excellent antibacterial and antifungal behavior of these compounds30. Another research work achieved in 20205 also confirmed their main antibacterial activity against a panel of gram-positive and harmful strains.
Despite their excellent antibacterial effects, in some cases, these pathogens strains can escape from the effect of these substances31,32. This new situation compelled the researchers to develop new promising strategies. Our study selected a simple method by combining the synthesized compounds and some antibiotics of references.
The approach adopted herein has given a satisfactory result. On the one hand, we noticed an increase in antibacterial activity. On the other hand, we obtained a powerful effect with a lower concentration of antibiotics.
CONCLUSIONS
In this study, four derivatives of α-amino phosphonates were successfully synthesized, then assessed for their antibacterial activity in vitro and silico.
The obtained data revealed that all studied compounds are biologically active and possess potent antibacterial activities, especially the 4a derivative.
With biological evaluation and docking results in hand, we can propose a plausible mode of action of these compounds before evaluating their biological activity in vitro and in vivo.
Limit of studies
Given the above facts, a single bioassay is not able to reflect the full picture of the derivatives' effects on multi-resistant bacteria
Recommendations for future studies
We recommend using another bacterial species (Gram- and Gram+) to provide broad information about the antibacterial potential of these compounds.
Acknowledgments
This work was supported by the CNRST (Moroccan research center of science and technology) under the Research Excellence Scholarship
Program.
Finding statement
This research was conducted within the university research plans and benefited from the general funds granted to the physiopathology, molecular genetics, and biotechnology laboratories.
Conflicts of Interest: None
Author contributions: B. Addoum, A. Soukri, B. El Khalfi, and H. Elmakssoudi contributed to the study's design. S. Harrati wrote and reviewed the draft of the manuscript.
REFERENCES
1. Vouga M, Greub G. Emerging bacterial pathogens: The past and beyond. Clin Microbiol Infect [Internet]. 2016;22(1):12–21. http://dx.doi.org/10.1016/j.cmi.2015.10.010
2. Bonneaud C, Weinert LA, Kuijper B. Understanding the emergence of bacterial pathogens in novel hosts. Philos Trans R Soc B Biol Sci. 2019;374(1782).
3. Kenawy ERS, Azaam MM, Saad-Allah KM. Synthesis and antimicrobial activity of α-aminophosphonates containing chitosan moiety. Arab J Chem. 2015 May 1;8(3):427–32.
4. Reddy GM, Garcia JR, Reddy VH, Kumari AK, Zyryanov G V., Yuvaraja G. An efficient and green approach: One pot, multi component, reusable catalyzed synthesis of pyranopyrazoles and investigation of biological assays. J Saudi Chem Soc. 2019 Mar 1;23(3):263–73.
5. Poola S, Nagaripati S, Tellamekala S, Chintha V, Kotha P, Yagani JR, et al. Green synthesis, antibacterial, antiviral and molecular docking studies of α-aminophosphonates. Synth Commun [Internet]. 2020;0(0):1–18. Available from: https://doi.org/10.1080/00397911.2020.1753079
6. Abdel-Megeed MF, Badr BE, Azaam MM, El-Hiti GA. Antimicrobial activities of a series of diphenyl (4-(aryldiazenyl)biphenyl- 4-ylamino)(pyridin-3-YL)methylphosphonates. Phosphorus, Sulfur Silicon Relat Elem. 2012 Oct 1;187(10):1202–7.
7. Addoum B, El Khalfi B, Daouda Mar P, Ansari NF, Elmakssoudi A, Soukri A, et al. Synthesis, Characterization of Some α-aminophosphonate Derivatives and Comparative Assessment of their Antioxidant Potential in-vivo and in-vitro. Vol. 2, Acta Scientific Pharmaceutical Sciences. 2018.
8. Rezaei Z, Firouzabadi H, Iranpoor N, Ghaderi A, Reza M. Design and one-pot synthesis of a -aminophosphonates and bis ( a -aminophosphonates ) by iron ( III ) chloride and cytotoxic activity. Eur J Med Chem. 2009;44:4266–75.
9. Subba G, Uma K, Rao M, Syama C, Suresh C. Neat synthesis and antioxidant activity of a -aminophosphonates. Arab J Chem [Internet]. 2014;7(5):833–8. Available from: http://dx.doi.org/10.1016/j.arabjc.2013.01.004
10. Sieńczyk M, Oleksyszyn J. Irreversible Inhibition of Serine Proteases - Design and In Vivo Activity of Diaryl alpha- Aminophosphonate ... Irreversible Inhibition of Serine Proteases – Design and In Vivo Activity of. Curr Med Chem. 2009;16:1673–87.
11. Aissa R, Guezane-Lakoud S, Kolodziej E, Toffano M, Aribi-Zouioueche L. Diastereoselective synthesis of bis(α-aminophosphonates) by lipase catalytic promiscuity. New J Chem. 2019;43(21):8153–9.
12. Rauhamäki S, Postila PA, Niinivehmas S, Kortet S, Schildt E, Pasanen M, et al. Structure-activity relationship analysis of 3-phenylcoumarin-based monoamine oxidase B inhibitors. Front Chem. 2018 Mar 1;6:3–18.
13. Bouchentouf S, Noureddine M. Identification of Compounds from Nigella Sativa as New Potential Inhibitors of 2019 Novel Coronasvirus ( Covid-19 ): Molecular Docking Study . ChemRxiv. 2020;(April).
14. Addoum B, El Khalfi B, Derdak R, Sakoui S, Elmakssoudi A, Soukri A. The one-pot synthesis of some bioactive pyranopyrazoles and evaluation of their protective behavior against extracellular H2O2 and SNP in T. Thermophila. Jordan J Biol Sci. 2021;14(1):31–9.
15. Zahouily M, Elmakssoudi A, Mezdar A, Rayadh A, Sebti S, Lazrek H. Three Components Coupling Catalysed by Na2CaP2O7: Synthesis of α-Amino Phosphonates Under Solvent-Free Conditions at Room Temperature. Lett Org Chem. 2005 Jul 9;2(5):428–32.
16. Zahouily M, Elmakssoudi A, Mezdar A, Rayadh A, Sebti S, Lazrek H. Three Components Coupling Catalysed by Na2CaP2O7: Synthesis of α-Amino Phosphonates Under Solvent-Free Conditions at Room Temperature. Lett Org Chem. 2005 Jul 9;2(5):428–32.
17. Jouimyi MR, Bounder G, Essaidi I, Boura H, Zerouali K, Lebrazi H, et al. Molecular docking of a set of flavonoid compounds with helicobacter pylori virulence factors CagA and VacA. J HerbMed Pharmacol. 2020;9(4):412–9.
18. da Silva FMA, da Silva KPA, de Oliveira LPM, Costa E V., Koolen HHF, Pinheiro MLB, et al. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the sars-cov-2 main protease (Mpro) and rna-dependent rna polymerase (rdrp). Mem Inst Oswaldo Cruz. 2020;115(9):1–8.
19. Ramesh DMK V. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. VirusDisease. 2020;2(31):1–6.
20. Filali OA, Soukri A, Khalfi B El. Evaluation of Antibacterial Effect of Some Essential Oils by Contact and Volatile Methods. 2019;9(4):81–90.
21. Kumar MH, Jones CS, Ravi A. Molecular docking studies of interaction between protease activated inhibitor receptor as the crucial protein and hydroxyl cinnamic acid as ligands. Asian J Pharm Pharmacol. 2019;5(1):137–42.
22. El Zoeiby A, Sanschagrin F, Levesque RC. Structure and function of the Mur enzymes: Development of novel inhibitors. Mol Microbiol. 2003;47(1):1–12.
23. Demeester KE, Liang H, Jensen MR, Jones ZS, D'Ambrosio EA, Scinto SL, et al. Synthesis of Functionalized N-Acetyl Muramic Acids to Probe Bacterial Cell Wall Recycling and Biosynthesis. J Am Chem Soc. 2018;140(30):9458–65.
24. Nadiveedhi MR, Nuthalapati P, Gundluru M, Yanamula MR, Kallimakula SV, Pasupuleti VR, et al. Green Synthesis, Antioxidant, and Plant Growth Regulatory Activities of Novel α-Furfuryl-2-alkylaminophosphonates. ACS Omega. 2021;6(4):2934–48.
25. Yang Y, Severin A, Chopra R, Krishnamurthy G, Singh G, Hu W, et al. 3,5-Dioxopyrazolidines, novel inhibitors of UDP-N- acetylenolpyruvylglucosamine reductase (MurB) with activity against gram-positive bacteria. Antimicrob Agents Chemother. 2006;50(2):556–64.
26. Yao G, Ye M, Huang R, Li Y, Pan Y, Xu Q, et al. Bioorganic & Medicinal Chemistry Letters Synthesis and antitumor activities of novel rhein a -aminophosphonates conjugates. Bioorg Med Chem Lett [Internet]. 2014;24(2):501–7. Available from: http://dx.doi.org/10.1016/j.bmcl.2013.12.030
27. Maleki B, Nasiri N, Tayebee R, Khojastehnezhad A, Akhlaghi HA. Green synthesis of tetrahydrobenzo[b] pyrans, pyrano[2,3-c] pyrazoles and spiro[indoline-3,4′-pyrano[2,3-c] pyrazoles catalyzed by nano-structured diphosphate in water. RSC Adv. 2016;6(82):79128–34.
28. Kunjapur AM, Tarasova Y, Prather KLJ. Synthesis and accumulation of aromatic aldehydes in an engineered strain of escherichia coli. J Am Chem Soc. 2014;136(33):11644–54.
29. Manolov I, Danchev ND. Synthesis, toxicological, and pharmacological assessment of some oximes and aldehyde condensation products of 4-hydroxycoumarin. Arch Pharm (Weinheim). 1999;332(7):243–8.
30. Rezaei Z, Khabnadideh S, Zomorodian K, Pakshir K, Nadali S, Mohtashami N, et al. Design , Synthesis , and Antifungal Activity of New $α$ -Aminophosphonates. 2011;2011.
31. Feleke T, Eshetie S, Dagnew M, Endris M, Abebe W, Tiruneh M. Multidrug ‑ resistant bacterial isolates from patients suspected of nosocomial infections at the University of Gondar Comprehensive Specialized Hospital , Northwest Ethiopia. BMC Res Notes [Internet]. 2018;1–7. Available from: https://doi.org/10.1186/s13104-018-3709-7
32. Li B, Webster and TJ. Bacteria antibiotic resistance: New challenges and opportunities for implant-associated orthopedic infections. J orthopeadic Res. 2018;36(1):22–32.
Received: 18 May 2021
Accepted: 12 July 2021
Bouchra El Khalfi1, Boutaina Addoum1, Suhayla Harrati1, Abdelhakim Elmakssoudi2 and Abdelaziz Soukri1, *
1Laboratory of Physiopathology, Genetics, Molecular and Biotechnology (PGMB), Department of Biology, Faculty of Sciences Aïn Chock, Research Center of Health and Biotechnology, Hassan II University of Casablanca, B.P 5366 Maarif, Casablanca, Morocco.
2 Laboratory of Organic Synthesis, Extraction, and Valorization, Department of Chemistry, Faculty of Sciences Aïn Chock, Hassan II University of Casablanca, B.P 5366 Maarif, Casablanca, Morocco.
Corresponding author: Addoum Boutaina,
PhD student,
Email address: [email protected]
Tel: +21266109549.
