2023.08.01.31
Files > Volume 8 > Vol 8 No 1 2023
Molecular characterization of national cocoa collection from the leading traditional growing areas in Ecuador
James Quiroz-Vera1, Eduardo Morillo2,* Carla Cordoba2,3 and Johana Buitron2,
1 Programa de Cacao. Instituto Nacional de Investigaciones Agropecuarias. Estación Experimental Litoral Sur; Yaguachi, Ecuador. Mail: [email protected] .
2 Departamento de Biotecnología. Instituto Nacional de Investigaciones Agropecuarias.Johana Buitron.
3 Estación Experimental Santa Catalina. Mejia,Ecuador
* Correspondence: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.01.31
ABSTRACT
Ecuador is the leading producer and exporter of fine cocoa, with plantations over 80 years old, preserving distinctive aroma and flavor characteristics. The research objective was to screen the genetic variability of a collection of National cocoa from Ecuador's leading traditional cocoa growing areas, denominated as Centennial National Cocoa Plants (CCNC). This germplasm collection with 243 accessions was analyzed with 20 microsatellites (SSR) markers. DNA genotyping was highly informative, generating a total of 109 SSR alleles with an average of 5.5 alleles per locus. Only 0.8% of duplicate accessions were identified. The average genetic diversity obtained was 0.447, and the polymorphic content index was 0.414, which shows a high genetic diversity. The clustering, main coordinates, and population assignment analysis revealed that the samples are classified into two subpopulations (GN and GM), differentiated by their level of heterozygosity, with a fixation index value of 0.105. The results showed that microsatellite markers and statistical tools provide useful information that favors managing and conserving genetic variability in CCNC collection.
Keywords: fine and aroma cocoa, Sabor Arriba, DNA genotyping, SSR markers
INTRODUCTION
Theobroma cacao L. is a fruit tree belonging to the genus Theobroma, corresponding to the Malvaceae family1. It is a diploid and allogamous species with a high degree of genetic diversity in its segregating populations2,3. Cocoa is an important crop that grows in tropical conditions, mainly in areas ranging from warm to humid, and on continents such as Asia, Africa, and the Americas. It is considered one of the world's most lucrative and commercialized products due to its organoleptic attributes4, 5.
The diversity of cocoa begins with the Criollo cocoa, followed by the Forastero cocoa, and finally, a kind of hybrid cocoa, a result of the mixture between these two kinds of cocoa called Trinitario, based on morphological traits of the crop6. Likewise, with more recent molecular data7, a new classification of cocoa types as proposed in 10 genetic populations called Marañón, Curaray, Criollo, Iquitos, Nanay, Contamana, Amelonado, Purús, Nacional (hereafter National), and Guyana.
In Ecuador, the first plantations of the National cocoa variety date back to the 1600s8, which were located along the shores of the Guayas River. Until the beginning of the 20th century, National cocoa was the only type of cocoa cultivated in Ecuador. From that time, there are still trees over 100 years old, called Centennial National Cocoa, which still retain the characteristics of fine cocoa and the aroma flavor. Genetic material was introduced between 1916 and 1919 to conserve the crop and reduce the diseases that affected the trees, which resulted in this type of cocoa disappearing from the production area and being replaced by hybrid materials, which nowadays present a high genetic variability9.
INIAP and the Tenguel Aroma Cocoa Center (CCAT) in Ecuador, in search of the rescue and preservation of these native National trees, established a collection of Centennial trees for study and utilization. Many plants were collected and preserved in ex-situ collections in its leading cocoa germplasm banks.
For the characterization of the cocoa germplasm collected, microsatellite markers or simple sequence repetition (SSR) are often used in cocoa. SSRs are the most commonly used markers in studies of plant genetic diversity, assignment of individuals to their population of origin, and determination of population structure, because they are very polymorphic and codominant, providing more genetic information than other types of markers 10. There is an excellent variety of cocoa-specific microsatellite markers with sequences previously described11 12,13, 14, and ones employed with molecular markers in National cocoa8, 15, 16, 17, 18, 19.
The study is part of a broader investigation into preserving Ecuador's National Centenary cocoa collection. It is highlighted that of the samples of trees over 100 years old, their genetic variability is unknown. These trees supposedly preserve their purity (homozygosity) and preserve distinctive characteristics of fine cocoa and aroma. Due to the above, the present study aimed to molecularly characterize a collection of Centenary National cocoa trees from Ecuador's main traditional cocoa growing areas, using a panel of 20 SSR markers.
MATERIALS AND METHODS
Biological material: A total of 243 plant samples were collected from the National Centenary cocoa collection (CCNC) of the South Coast Experimental Station of INIAP, located in the Yaguachi canton of the Guayas province. Each sample was coded depending on the origin of the trees from which they were taken: "M" for samples taken from trees from the region of Manabí and "Lr" for those that were taken from trees from the province of Los Rios.
DNA genotyping: DNA was extracted from cocoa leaf tissue by spectrophotometry and stored at -20°C 20, 21. For SSR analysis, 20 cocoa-specific genomic microsatellite markers were used. The forward primers were marked with fluorescent dyes (M13 tailing), and the PCR products were separated by vertical electrophoresis in the LICOR-4300 equipment 22. The allelic profile obtained was visualized in the SAGAGT-SSR™ program (LI-COR Biosciences), where the genotyping was performed, and a genotypic matrix was obtained that listed the size of the alleles of each sample for each marker.
Identification of representative accessions: Duplicate samples were identified by pairwise comparisons among the 243 samples based on their available alleles reported in their allelic profile, using the Microsatellite Toolkit program 23. From the refined genotypic matrix, samples were identified that presented a single allele for at least 16 microsatellite markers of the 20 used, which had a high level of homozygosity (≥80%). These samples are representative since, having this level of homozygosity, they are considered pure samples, and it is estimated that they retain characteristics of fine National cocoa and aroma.
Genetic diversity analysis: The study of the entire population was performed using the PowerMarker v3.25 program 24. Several statistical parameters were determined, such as the effective number of alleles, allelic frequencies, genotypic frequencies, observed heterozygosity (Ho), expected heterozygosity (He), and the polymorphic information content (PIC)25. Using the same program, a bootstrap analysis of 999 permutations and 100 repetitions was performed. These data were used in the PHYLIP 3.67 program to generate a consensus tree. Bayesian clustering analysis using Structure v.2.3.4 software was applied to determine the population structure, with K values from 1 to 6, with a Burnin period of 50000, a Marko v Chain Monte Carlo (MCMC) value of 50000 with 10 simulations. The Structure Harvester software was also used to establish the maximum value of ∆k 26. The pairwise distances were indicated in a Principal Coordinate Analysis (PcoA). With the formed subpopulations, a molecular analysis of variance (ANOVA) was performed, and the F statistics, Fis (intrapopulation inbreeding index), Fit (total inbreeding index), and Fst (fixation index) were established using the software GenAlex v6.5 27.
RESULTS
Identification of duplicates
In the pairwise comparison based on the SSR multilocus profile, only two duplicate accessions were identified within the CCNC collection. These two samples shared 38 alleles. Total duplication represents 0.8% of genotyped samples from the CCNC collection.
Identification of representative samples
Thirteen samples were identified as representative samples within the CCNC collection. These samples are those that presented high levels of homozygosity.
Population structure analysis
The population structure simulation established a ∆k value equal to 2; that is, the population was divided into two main clusters or subpopulations (Fig. 1). From the Q index, 173 samples were assigned to one of these two subpopulations with a high level of probability (Q index 0.9-1), 99 samples belonging to the subpopulation identified in green color and 74 samples belonging to the subpopulation identified in red. Within the 99 samples grouped in the green subpopulation, 86 presented a homozygosity level of less than 80%, and the remaining 13 samples presented a high level of homozygosity (≥80%), which is why this group was called GN (National Group), and the subpopulation identified in red, made up of the 74 samples, was called GM, referring to the subpopulation of hybrid samples.
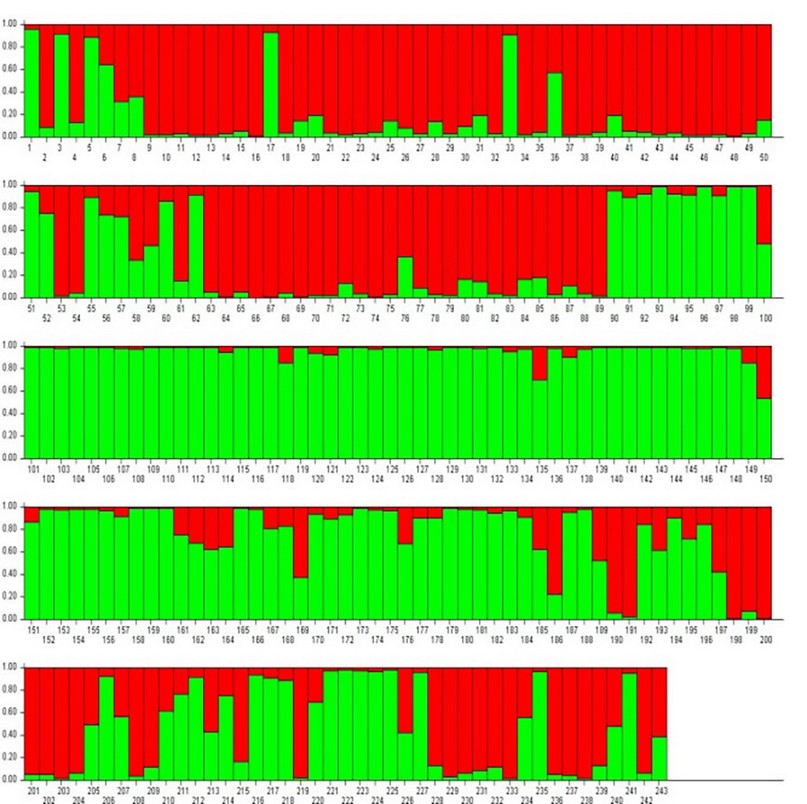
Figure 1. Bayesian population assignment using STRUCTURE (K=2)
This population organization was identified in the PcoA analysis (Fig. 2). The GN subpopulation is more homogeneous since the samples present a high genetic similarity; the GM group is much more diverse since it offers more significant genetic divergence between the samples.
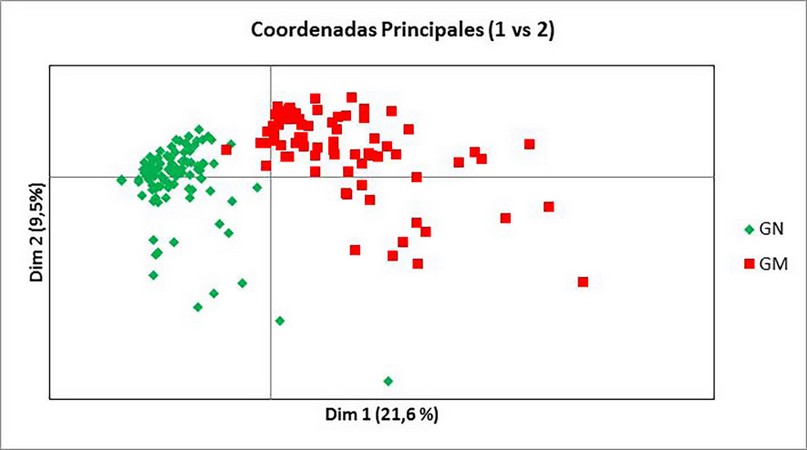
Figure 2. PcoA graph of the 173 accessions assigned to one of the two subpopulations (first coordinate = 18.1% of the total, and the second = 6.1)
Genetic diversity analysis
A total of 109 alleles were identified for the entire population, with a mean of 5.5 alleles per locus. The mean genetic diversity (He) was 0.447, the observed heterozygosity (Ho) was 0.331, and the polymorphic information index (PIC) was 0.414. The GN subpopulation presented a mean genetic diversity of 29.5%, and the GM subpopulation of 55.4%. Thirty-six exclusive alleles for the GM subpopulation were also identified. The results of the ANOVA and the F statistics for the two subpopulations determined that there is a variability of 70% (Fis=0.178) within the subpopulations and a variability of 30% (Fst=0.105) between the subpopulations.
DISCUSSION
Molecular markers have proven adequate for characterizing genetic variability in T. cocoa 17, 18, 28, 29, 30, 31, 32, 33. In the present study, samples from a cocoa collection called National Centenarian cocoa collection (NCCC) were used, which is made up of 260 accessions collected from farms where trees with characteristics of National cocoa were found and whose ages were 75-100 years, located in the northern area of the province of Manabí, and in the province of Los Rios in Ecuador.
In a general context, the CCNC collection presented considerable genetic diversity, taking into account both the level of He and the fact that it was found only one case of duplication among the accessions. This corresponds to 0.8%, much lower than the data reported in other studies, such as 19.6% of duplicates 28, 9.1% 29, and 12.9% 34.
It was shown that CCNC genetic diversity is structured into two groups or subpopulations: the GN and the GM groups. Similar results were reported in previous studies15 by grouping, and a dendrogram identified two clusters or subpopulations in a population of National cocoa obtained from plantations that were 80 to 100 years old. These two subpopulations showed significant differences based on their average level of heterozygosity; the first subpopulation is characterized by being more homogeneous since it only includes samples of National cocoa and a low level of heterozygosity (5%), and the second presents a high level of heterozygosity (44%) because it is more heterogeneous and includes samples of National cocoa trees from Venezuelan and hybrid models.
Furthermore, results similar to those reported by other researchers were evidenced, showing that Ecuadorian cocoa is divided into two subpopulations16: a subpopulation with a low level of heterozygosity (5%), considered to be representative of National cocoa; and a subpopulation with a higher level of heterozygosity (32%), considered to be representative of modern National cocoa, made up of hybrid samples. Previous researchers established that a large part of modern National cocoa cultivation corresponds to the so-called National Trinitarian complex35, formed from the introduction of Trinitarian-type Venezuelan cocoa at the end of the 19th century and beginning of the 20th century and its subsequent gene flow with the National Centennial cocoa population. It was also established that the high homozygosity of several Ecuadorian cocoa samples may be due to the self-compatibility of the oldest National cocoa from Ecuador8 and that the compatibility variation of the modern National cocoa is also due to the introgression of the Trinitario cocoa genome. It can be inferred that the GN subpopulation group samples of the purest National type present less gene introgression from other cocoa varieties. On the other hand, the GM subpopulation includes the accessions with the highest level of heterozygosity. The percentage of heterozygosity presented by the GN subpopulation is 29.5%, which is considerably high compared to those published by other researchers15,16; this is because only 13 samples belonging to this subpopulation presented a high level of homozygosity. The remaining 86 genotypes did not present the same level of homozygosity but rather a genetic difference with the samples of the GM subpopulation. This genetic differentiation allows these two subpopulations to appear separate from one another in the multivariate PCoA analysis.
The level of genetic diversity of the GM subpopulation was also evidenced due to the presence of the 36 exclusive alleles. It is inferred that the level of heterozygosity of this subpopulation could be the effect of the introgression of genes from various cocoa varieties, not only from Trinitarian cocoa 8. However, further studies are needed to establish the origin of this genetic diversity. The results obtained in this research are essential and valuable to continue characterization studies in Ecuador cocoa crops since they allow the identification of National cocoa trees with a high level of homozygosity. In addition, the results make it possible to establish genetic improvement programs to recover the characteristics of delicate and aroma cocoa that distinguish Ecuadorian National cocoa as the best worldwide. Improving the quality of this type of cocoa will allow it to be reactivated and commercialized in the international market, giving a new impetus to the country's economy.
CONCLUSIONS
We showed the efficiency of the panel of SSR markers used here in the genetic assignment of accessions. The CCNC collection presented a high genetic diversity with little duplication. Genetic diversity was identified in two groups. Firstly, the GN subpopulation includes 99 samples, of which 13 presented homozygosity ≥ 80%; hence they are considered samples of native National cocoa that still conserve their characteristics of fine cocoa and aroma. Likewise, the GM subpopulation includes 74 samples with high diversity, with 36 exclusive alleles, probably due to the introgression of genes from other cocoa varieties.
Author Contributions: Conceptualization, JQ and EM; methodology, EM, JB, CC; software, EM and JB; validation, JB; formal analysis, CC and JB; initial draft preparation, CC and JQ; writing—review and editing, EM and JB; funding acquisition, JQ. All authors have read and agreed to the published version of the manuscript. Please turn to the CRediT taxonomy for the term explanation. Authorship must be limited to those who have contributed substantially to work reported.
Funding: This research was funded by INIAP and GIZ
Acknowledgments: Authors thank the technical support from MAG and INIAP for germplasm establishment and other materials used for this project.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
1. Ballesteros William. Caracterización morfológica de árboles elite de cacao (Theobroma cacao L.) en el municipio de Tumaco, Nariño, Colombia. 2011. Tesis Doctoral. Universidad de Nariño.
2. Argout X, Salse J, Aury J, Guiltinan M, Droc G, Gouzy J, Allegre M, Chaparro C, Legavre T, Maximova S, Abrouk M, Murat F, Fouet O, Poulain J, Ruiz M. Roguet Y, Rodier-Goud M, Barbosa-Neto J. F, Sabot F, Lanaud C. The genome of Theobroma cacao. Nature Genetics. 2011; 43(2), 101–108.
3. López-Hernández, J. A., Ortiz-Mejía, F. N., Parada-Berríos, F. A., Lara-Ascencio, F., & Vásquez-Osegueda, E. A. Caracterización morfoagronómica de cacao criollo (Theobroma cacao L.) y su incidencia en la selección de germoplasma promisorio en áreas de presencia natural en El Salvador. Revista Científica Multidisciplinaria de la Universidad de El Salvador-Revista Minerva, 2019, vol. 2, no 1, p. 31-50.
4. Samaniego, I., Espín, S., Quiroz, J., Rosales, C., Carrillo, W., Mena, P., & García-Viguera, C. Effect of the growing area on the fat content and the fatty acid composition of Ecuadorian cocoa beans. International Journal of Food Sciences and Nutrition. 2021; 72(7), 901-911.
5. Samaniego, I., Espín, S., Quiroz, J., Ortiz, B., Carrillo, W., García-Viguera, C., & Mena, P. Effect of the growing area on the methylxanthines and flavan-3-ols content in cocoa beans from Ecuador. Journal of Food Composition and Analysis, 2020; 88, 103448.
6. Cheesman E. Notes on the nomenclature, classification and possible relationships of cocoa populations. Tropical Agriculture. 1944; 21, 144–159.
7. Motamayor J. C, Lachenaud P, da Silva e Mota J, Loor R, Kuhn D, Brown J, Schnell R. Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L). PLoS ONE. 2008; 3(10).
8. Loor R, Risterucci A, Courtois B, Fouet O, Jeanneau M, Rosenquist E, Amores F, Vasco A, Medina M, Lanaud C. Tracing the native ancestors of the modern Theobroma cacao L. population in Ecuador. Tree Genetics and Genomes. 2009; 5(3), 421–433.
9. Quiroz J, Morillo E, Samaniego I. Estudio de la expresión y diversidad genética para la determinación de la calidad en clones de cacao Nacional centenario del INIAP. 2019. No publicado.
10. Garrido-Cardenas J, Mesa-Valle C, Manzano-Agugliaro F. Trends in plant research using molecular markers. In Planta. 2018; 247(3), 543–557.
11. Lanaud C, Risterucci A, Pieretti I, Falque M, Bouet A, Lagoda P. Isolation and characterization of microsatellites in Theobroma cacao L. Molecular Ecology. 1999; (8), 2141–2152.
12. Risterucci A, Grivet L, N´ Goran J, Pieretti I, Flament M, Lanaud C. A high-density linkage map of Theobroma cacao L. Theoretical and Applied Genetics. 2000; (101), 948–955.
13. Saunders J, Mischke S, Leamy E, Hemeida A. Selection of international molecular standards for DNA fingerprinting of Theobroma cacao. Theoretical and Applied Genetics. 2004; 110(1), 41–47.
14. Pugh T, Fouet O, Risterucci A, Brottier P, Abouladze M, Deletrez C, Courtois B, Clement D, Larmande P, N’Goran J, Lanaud C. A new cacao linkage map based on codominant markers: Development and integration of 201 new microsatellite markers. Theoretical and Applied Genetics. 2004;108(6), 1151–1161.
15. Lerceteau E, Quiroz J, Soria J, Flipo S, Pétiard V, Crouzilat D. Genetic differentiation among Ecuadorian Theobroma cacao L. accessions using DNA and morphological analyses. In Euphytic. 1997; Vol. 95.
16. Crouzillat D, Bellanger L, Rigoreau M, Bucheli P, Pétiard V. Genetic structure, characterisation and selection of nacional cocoa compared to other genetic groups. In F. L. Bekele (Ed.), Proceedings of the international workshop on new technologies and cocoa breeding. INGENIC. 2001.
17. Quiroz J. Caracterización Molecular y Morfológica de Genotipos Superiores con Caracteristicas de Cacao Nacional (Theobroma cacao L.) de Ecuador. Centro Agronómico Tropical de Investigación y Enseñanza. 2002.
18. Loor Rey Gastón. Caracterización Morfológica y Molecular de 37 clones de cacao (Theobroma cacao L.) Nacional de Ecuador 2002. 108 p. INIAP Archivo Historico.
19. Loor-Solorzano R, Fouet O, Lemainque A, Pavek S, Boccara M, Argout X, Amores F, Courtois B, Risterucci A, Lanaud C. Insight into the Wild Origin, Migration and Domestication History of the Fine Flavour Nacional Theobroma cacao L. Variety from Ecuador. PLoS ONE. 2012; 7(11).
20. Doyle J, Doyle J. Rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987; 19(1), 11–15.
21. Russell A, Samuel R, Rupp B, Barfuss M, Šafran M, Besendorfer V, Chase M, Safran M, Besendorfer V, Chase M. Phylogenetics and cytology of a pantropical orchid genus Polystachya (Polystachyinae, Vandeae, Orchidaceae): Evidence from plastid DNA sequence data. In Source: Taxon. 2010; Vol. 59, Issue 2.
22. Morillo E, Miño G. Marcadores Moleculares en Biotecnología Agrícola: Manual de procedimientos y técnicas en INIAP. Manual No. 91. Instituto Nacional Autónomo de Investigaciones Agropecuarias, Estación Experimental Santa Catalina. 2011; 121 p.
23. Kim K, Sappington T. Microsatellite Data Analysis for Population Genetics. 2013; 271–295.
24. Liu K, Muse S. PowerMaker: An integrated analysis environment for genetic marker analysis. Bioinformatics. 2005; 21(9), 2128–2129.
25. Caruso G, Broglia V, Pocovi M. Diversidad genética. Importancia y aplicaciones en el mejoramiento vegetal. 2015; 4(1), 45–50.
26. Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology. 2005; 14(8), 2611–2620.
27. Peakall R, Smouse P. GenALEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012; 28(19), 2537–2539.
28. Zhang D, Mischke S, Johnson E, Phillips-Mora W, Meinhardt L. Molecular characterization of an international cacao collection using microsatellite markers. Tree Genetics and Genomes. 2008; 5(1), 1–10.
29. Irish B, Goenaga R, Zhang D, Schnell R, Brown J, Motamayor J. Microsatellite fingerprinting of the USDA-ARS tropical agriculture research station cacao (Theobroma cacao L.) Germplasm collection. Crop Science. 2010; 50(2), 656–667.
30. Romero C, Bonilla J, Santos E, Peralta E. Identificación Varietal de 41 Plantas Seleccionadas de Cacao (Theobroma cacao L.) Provenientes de Cuatro Cultivares Distintos de la Región Amazónica Ecuatoriana, Mediante el Uso de Marcadores Microsatélites. Revista Tecnológica ESPOL. 2010; 23, 121–128.
31. Lozada-Vargas P. Caracterización molecular de 42 accesiones de la colección de genotipos de cacao Nacional (Theobroma cacao L.) de la EET PICHILINGUE, INIAP, mediante el uso de marcadores microsatélites (SSRs). ESPE. 2014.
32. López-Gómez, P., Avendaño-Arrazate, C. H., Iracheta-Donjuan, L., & del Carmen Ojeda-Zacarías, M.. PCR-SRAP/ITAP para la caracterización molecular del género Theobroma. Revista Fitotecnia Mexicana. 2021; 44(1), 3-3.
33. Ricaño-Rodríguez J, Hipólito-Romero E, Ramos-Prado J, Cocoletzi-Vásquez E. Genotyping-by-Sequencing of native varieties of Theobroma cacao (Malvaceae) from the States of Tabasco and Chiapas, Mexico. Botanical Sciences. 2019; 97 (3): 381-397.
34. Rosero C. Selección estable de marcadores moleculares microsatélites (SSRs) para la identificación de clones comerciales de cacao Nacional (Theobroma cacao L.), recomendados por el INIAP. ESPE. 2013.
Amores F, Palacios A, Jiménez J, Zhang D. Entorno ambiental, genética, atributos de calidad y singularización del cacao en el Nor Oriente de la provincia de Esmeraldas. Boletín Técnica. 2009, N°35.
Received: October 23, 2022 / Accepted: January 15, 2023 / Published:15 February 2023
Citation: Quiroz-Vera J, Morillo E, Cordoba C, Buitron J. Molecular characterization of national cocoa collection from the leading traditional growing areas in Ecuador. Revis Bionatura 2023;8 (1)31. http://dx.doi.org/10.21931/RB/2023.08.01.31
