2023.08.03.27
Files > Volume 8 > Vol 8 No 3 2023
Preview / Index / Next
Quantitative and Qualitative Determination of Biofilm Formation Pattern in Multidrug Resistance Acinetobacter baumannii in Correlation with COVID-19 and Respiratory Infected Patients

1, 3 National Center for Educational Laboratories, Medical City, Ministry of Health Baghdad, Iraq.
2Al-Iraqi University, College of Medicine, Medical Microbiology Department, Baghdad, Iraq
Available from: http://dx.doi.org/10.21931/RB/2023.08.03.27
ABSTRACT
Respiratory tract infection (RTI) is one of the triggering factors that cause severe and life-threatening health problems. Acinetobacter baumannii frequently causes nosocomial infections, especially in intensive care units (ICUs). Multidrug-resistant (MDR) A. baumannii encodes multiple virulence factors that contribute to chronic diseases, as well as antibiotic resistance; among them is the capacity to colonize and produce Biofilm on biotic and abiotic surfaces that is the primary source of worry in hospital environments. The study assessed the role of antibiotic resistance and biofilm formation patterns caused by MDR A. baumannii between isolates from COVID-19 and respiratory-infected patients in Baghdad Medical City hospitals and Teaching laboratories. This study included isolating and identifying A. baumannii from the sputum samples of respiratory tract-infected patients. (some of them infected with COVID-19) . Diagnosis of isolates was done by conventional cultural, microscopical and biochemical methods and confirmed by API 20E and VITEK2 system. Investigation of the susceptibility profile of the isolates against 18 types of antibiotics was tested, and the determination of A. baumannii biofilm formation was recorded by qualitative and quantitative methods. Results showed that 36 bacterial isolates were identified as A. baumannii. Most isolates were resistant to antibiotics, including all penicillins and cephalosporins (including inhibitor combinations), fluoroquinolones and aminoglycosides, followed by carbapenems antibiotics.
In contrast, all isolates were susceptible to colistin (94.4%), followed by minocycline (36.1%), and the proportions of MDR, XDR, and PDR were 19.4%, 77.8 %, and 2.8 %, respectively. All isolates of A. baumannii showed biofilm formation (100%) by Quantitative method(MTP) and 91.6%. By the qualitative approach, we can conclude that there were no significant changes in biofilm values after 48 hours between groups with and without COVID-19 infection. However, there was a significant difference in adherence levels of bacteria between COVID-19 groups, with a higher proportion of solid adherence in the COVID-19 group compared to moderate adherence in the COVID-19-free group patients.
Keywords: Acinetobacter baumannii, Antibiotic Susceptibility, Biofilm formation patterns, COVID-19 and respiratory infected patients.
INTRODUCTION
Infectious disorders that affect the respiratory system are widespread throughout the world. The common cold, influenza, tuberculosis, and pneumonia are separate from those listed here. 1 Many different viral infections can cause disease in the upper respiratory system. The common cold and the flu are only two examples of these viruses. Bacterial infections in the lower respiratory tract, such as tuberculosis and pneumonia, are a leading cause of severe illness and can even be deadly in some cases. 2 Nosocomial pathogen infections are secondary infections commonly caused by pathogens spread in hospitals. Hospitals are a significant source of these pathogens. 3. In hospitalized patients, one of the most prevalent causes of infection caused by Gram-negative bacteria (GNB), Acinetobacter baumannii, is one of the most common causes. A. baumannii has been linked to a higher death rate and antibiotic resistance4,5. One of the key factors contributing to chronic and persistent infections and antibiotic resistance is the ability of A. baumannii to colonize and form Biofilm on biotic and abiotic surfaces6. Biofilms are communities of microorganisms attached to biotic and abiotic surfaces and encased in a matrix of extracellular polymeric substances (EPS). In terms of their physiology, they are distinct from bacteria that float freely in the water 7. Because biofilm-encased cells have low metabolic activity and are shielded by the extracellular matrix, They will be more resistant to medicines and the host's innate immune system 8. Biofilm infections, notably ventilator-associated pneumonia, are caused by A. baumannii. 9 MDR A. baumannii (MDR-AB) infections are a significant concern in patients admitted to the intensive care unit10. Poor outcomes are linked to this infection, associated with high fatality rates, particularly in intensive care unit (ICU) patients. Inappropriate therapy and restricted therapeutic alternatives contribute to poor outcomes 11,12. Coronavirus Disease-2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has been spread in the entire world13. Acute respiratory failure is a medical emergency requiring the patient to be treated at the highest possible level, provided by the intensive care unit of the hospital (ICU). More than ten percent of these individuals will require some form of mechanical ventilation, either noninvasive or invasive 14. These patients are administered broad-spectrum antibiotics as a precautionary measure, which increases the likelihood that they will contract an illness caused by multidrug-resistant (MDR) germs15. During the COVID-19 pandemic, A. baumannii coinfection with SARS-CoV-2 was recorded in several locations, including Wuhan (China), France, Spain, Iran, Egypt, New York (USA), Italy, and Brazil. These are only a few of the locations 16. The Acinetobacter spp. Bacteria are the second most prevalent form of bacteria detected in patients with positive bacterial cultures, and they are the third major cause of death in patients infected with COVID-19 and another virus at the same time 17. This research aimed to examine, quantitatively and qualitatively, the biofilm formation patterns of different isolates of multidrug-resistant Acinetobacter baumannii collected from people with respiratory illnesses brought on by the COVID-19 virus.
METHODS
Samples collection
Two hundred sputum samples were collected from respiratory infection patients (15 of them from COVID-19 patients admitted to ICU) in Medical City Hospitals in Baghdad, Iraq, with ages ranging between 1 to 80 years old, from--November 2021 -to February 2022. Sputum samples were put in sterile, wide-mouthed containers and immediately transported to the laboratory for further investigation.
Isolation and Identification of Bacteria
In the laboratory (Teaching Laboratories/Medical City, Baghdad), the collected specimens were cultured directly on blood agar and MacConkey agar and then incubated under aerobic conditions for 24 hrs at 37°C18. After the incubation period, bacteria were diagnosed by examining the macroscopic features of bacteria cells, Gram staining, and Biochemical tests, including Oxidase test, Catalase test, Urease test, Indole test, Citrate Utilization test, Kligler Iron Agar test, Motility test, Lactose fermentation on MacConkey agar, growth at 44 °C test, and Hemolysis on blood agar 18. A. baumannii was confirmed by the API-20E and VITEK 2 system.
Antibiotic sensitivity test
A Kirby-Bauer was applied to test the antibiotic susceptibility test was conducted 19. The outcomes were interpreted following Clinical Laboratory Standard Institute recommendations 20. The 18 antibiotic discs utilized in this investigation were. Amikacin (AK, 30μg), Tobramycin(TN,10 μg), Gentamicin(GN,10 μg) Cefotaxime (CTX, 30μg), Ceftazidime (CTZ, 30μg), Ceftriaxone (CTR, 30μg), Ciprofloxacin (CIP, 5μg),Levofloxacin(LEV,5 μg), Trimethoprim-Suphamethoxazole (TS, 1.25/23.75μg) ,Ticarcillin-clavulanate(TIM,75/10 μg), Ampicillin-Sulbactam(SAM,10/10 μg) Tetracycllin(T,30 μg),Doxycyclin(DXT,30 μg), ) Augmentin (AMG, 20 μg) ,Tobramycin(TN,10 μg) ,Imipenem(IMI ,10 μg), Meropenem (MEM,10 μg), Piperacillin(PRL,100 μg) and Piperacillin-Tazobactam(PTZ,100/10 μg). These discs were provided by (Bioanalyse, Turkey). The antibiotic tests were duplicated and quality controlled using Pseudomonas aeruginosa ATCC 27853 (incubated at 37°C for 18-24.). The resistant isolates to at least three antimicrobial classes are considered MDR. Another measurement of antibiotic sensitivity by Minimum Inhibitory Concentration (MIC) was made by VITEK 2 system.
Biofilm Formation
To evaluate the development of biofilm formation in A.baumannii isolates, two methods were used as follows:
Qualitative method (Congo red agar)
Study biofilm development in bacterial cultures by Congo-Red Agar Method (CRA). Black and dry crystalline colonies indicated a positive, strong isolation. The absence of a dry crystalline and the darkness of the colonies showed intermediate results. Weak biofilm formation was pink, with black areas in the core of colonies. 21
Quantitative method (Microtiter plate assay)
According to the method described by 22. the Biofilm Formation Microtiter Plate Assay was performed. isolates were grown in Luria-Bertani broth contain( 0.25) percent glucose overnight at 37 °C (LBG). In freshly made LBG pre-warmed to 37 °C, the culture was diluted 1:50. The sterile 96-well polystyrene microtitre plates were then incubated for 48 hours at 37 °C with the suspension as the inoculum. Any leftover biofilm was stained with crystal violet 1 percent (w/v) for 30 minutes and then rinsed with phosphate buffer saline (PBS) three times. Using 200 microliter ethanol/acetone (80:20 v/v) to resolubilize the dye attached to the adherent cells, the ELISA reader was used to measure the optical density O.D570 (Synergy4; BioTek). Three readings for each test were performed in duplicate for each. The evaluated strains' adhesion capacities were divided into four groups, and the cutoff optical density was established based on the average optical density of the negative control (which contained broth) (ODc). The following is how the results were interpreted:
-If OD ≤ ODc, the bacteria were non-adherent.
-If ODc < OD ≤ 2×ODc, the bacteria were weakly adherent.
-If 2×ODc < OD ≤ 4×ODc, the bacteria were moderately adherent.
-If 4×ODc < OD, the bacteria were strongly adherent.
RESULTS
Isolation and identification of bacteria
Results are shown in figure (1), which represents the growth of isolated bacteria on the surfaces of MaCconkey agar (A), which appear as small pink to lavender color colonies in a non-lactose fermenter. (B) represents the growth of isolates on blood agar media; colonies appear translucent to opaque, non-hemolytic and pigmented. All isolates appeared gram-negative, coccbacili under the light microscope 40 X, and also can grow at 44⁰C.

Figure 1. A.baumannii on (A) MacConky agar and (B) blood agar media
For biochemical tests, results show in Table (1) that all isolates gave negative consequences for the Indole test, oxidase test, and urease, and positive outcomes for catalase and utilization of citrate. The killer was developed to alkaline.

Table 1. Biochemical tests of A. baumannii
After the confirmation of the diagnosis by the API-20 E system and by the VITEK2 system, results showed that from the two hundred sputum samples, 36 samples were identified as A. baumannii bacteria, which were isolated from a respiratory infection. The age of patients ranged from 1 to 80 years old. The ratio of females to males was 11(30.6)and 25 (69.4), respectively. However, about one-fourth of the study sample had positive COVID-19 infection (22.2%) (8 isolates from 15 COVID-19 patients were identified as A. baumannii.
Antibiotic sensitivity test
Figure (2) shows a high resistance level of A. baumannii clinical isolates to most tested antibiotics using the Kirby-Bauer method. The present study revealed that all A. baumannii clinical isolates had 100% resistance to piperacillin-tazobactam, piperacillin, trimethoprim-sulfamethoxazole, augmentin, cefotaxime, ceftriaxone, ceftazidime, and ciprofloxacin. The 97.2% to ampicillin-sulbactam, ticarcillin-clavulanate, gentamicin, Amikacin, and levofloxacin, followed by Meropenem 94.4%, Tobramycin 86.1%, tetracycline 80.6%, and doxycycline 58.3%. A higher intermediate resistance was identified by bacteria against imipenem 19.4%; the VITEK 2 system did another measurement of antibiotic MIC. The rates of resistance were shown in Figure (3), with almost similar results shown in the Kirby-Bauer method; results also found that the bacteria revealed a higher sensitivity for colistin (94.4%) respectively, followed by minocycline (36.1%).
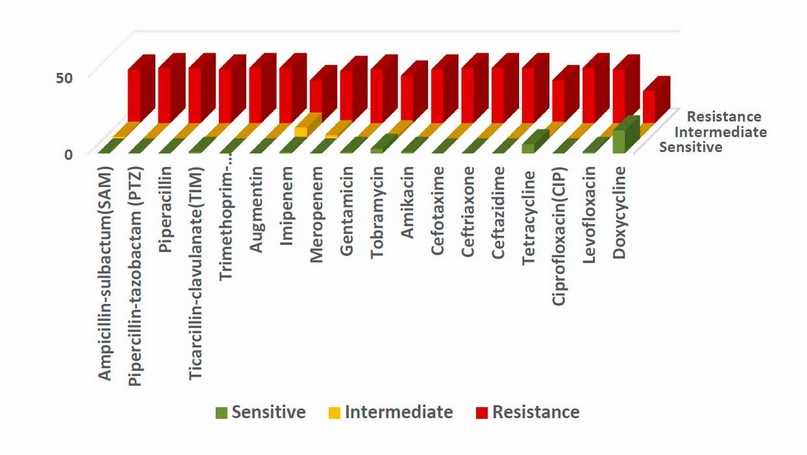
Figure 2. Antibiotic resistance of 36 A. baumannii clinical isolates by Kirby-Bauer method
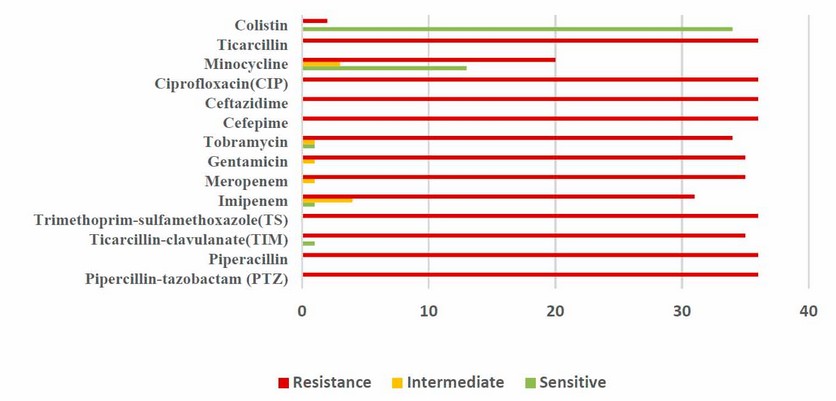
Figure 3. MIC antibiotic susceptibility of A. baumannii isolates by VITEK 2 system
Furthermore, according to the categorized classification of antibiotic resistance, A. baumannii revealed a higher proportion of Extensively-Resistance (28; 77.8%), followed by Multidrug Drug-Resistance (7; 19.4%) and solely (1; 2.8%) revealed to have Pan Drug-Resistant bacteria—figure (4).

Figure 4. Classification of antibiotic susceptibility of isolated A. baumannii bacteria (n=36)
Detection of the Bacterial Ability for Biofilm Formation:
The ability of 36 isolates of A. baumannii to adhere and to produce a slime layer (Biofilm formation) was experienced quantitatively by Microtiter plate (MTP) using an ELISA reader and qualitatively by using Congo-red agar (CRA). The currently obtained results showed in Table (2), Figure (5) and (6) that all isolates were biofilm producers by microtiter plate (MTP), 63.8%, 36.1% moderate and robust biofilm producers, respectively, after 48 hours incubation and the mean value of Biofilm after 48 hours (0.52367 ± 0.080312) was higher than that of the substantial cutoff value of 0.488 (OD of 0.122 x 4) with significant differences of 0.035667 (t= 2.665, df: 35, P= 0.012).

Figure 5. Biofilm formation of Acinetobacter baumannii on the microtiter plate.
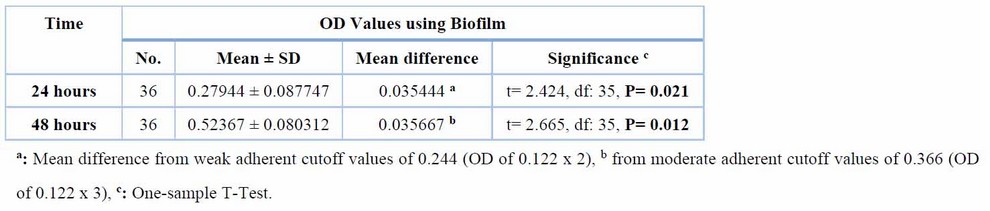
Table 2,Adherent characteristics of A.baumannii in 24 and 48 hours within the Biofilm

Figure 6. The OD adherent values of Acinetobacter baumannii in 24 and 48 hours within Biofilm (n=36)
Furthermore, there are no significant changes in mean biofilm values after 48 hours between groups with and without COVID-19 infection (0.55625 0.062089 vs. 0.51436 0.083409) (t= -1.315, df: 34, P= 0.197) Table (3):
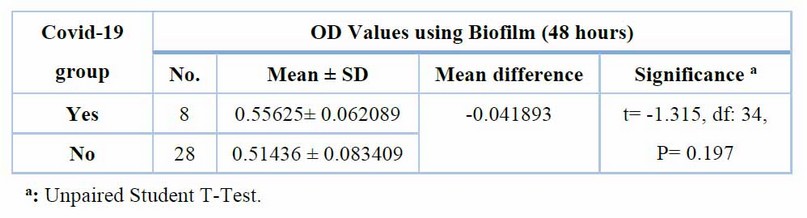
Table 3 Adherent characteristics of A. baumannii after 48 hours within Biofilm among covid-19 groups (n=36)
Furthermore, using Congo red, the adherent features of bacteria revealed that (36.1 % )of bacteria had a strong adherence characteristic after 24 hours, which grew gradually after 48 hours to (47.2 %) from moderate adherences (55.6 % and 44.4 %, respectively). In Congo red agar, however, 8.3% of bacteria showed no adherence after 24 or 48 hours, respectively, as shown in the Figure. (7,8)
However, there was a significant difference in adherence levels of bacteria using Congo red between covid-19 groups, with a higher proportion of solid adherence in the COVID-19 group compared to moderate adherence in the covid-19 free group (87.2 % vs. 53.6 ) (Fisher Exact test: 5.869, P = 0.048) table (4).

Figure 7. Black pigment of A. baumannii within Congo red agar (n=36)
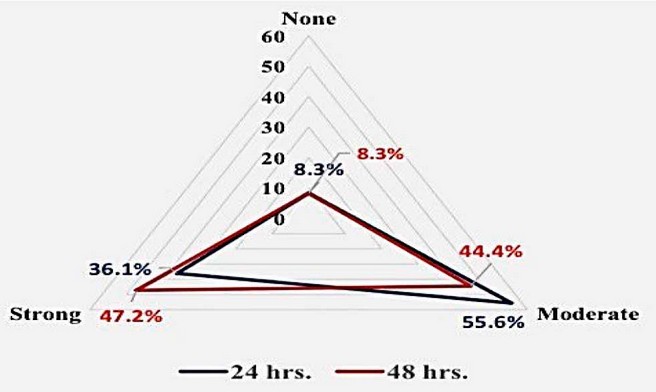
Figure 8. The distribution of adherent levels of Acinetobacter baumannii after 24 and 48 hours of incubation in Congo red agar (n=36)

Table 4. The distribution of adherent levels of Acinetobacter baumannii after 48 hours using Congo red agar according to covid-19 groups (n=36)
DISCUSSION
The 36 A.baumannii isolates had 100% resistance to cefotaxime, ceftriaxone, ceftazidime, piperacillin-tazobactam, piperacillin, trimethoprim-sulfamethoxazole, and ciprofloxacin. The 97.2% to ampicillin-sulbactam, ticarcillin-clavulanate, gentamicin, Amikacin, and levofloxacin, followed by Meropenem 94.4%, Tobramycin 86.1%, tetracycline 80.6%, and doxycycline 58.3%.Moreover, A higher intermediate resistance was identified by bacteria against imipenem 19.4%, and a higher sensitivity and lower resistance for colistin (94.4% vs. 5.6%), respectively, followed by minocycline(36.1%). Many studies found that A.baumannii has a high resistance to a broad spectrum of antibiotics; the results of our study agree with those mentioned by 23. Erbil City hospitals recorded A. baumannii isolates were resistant (100%) to cefotaxime, ciprofloxacin, levofloxacin, Amikacin, gentamicin, piperacillin, piperacillin-tazobactam trimethoprim-sulfamethoxazole, imipenem and meropenem), Followed by (98.2%) of the isolates were resistant to Ceftazidime and Cefepime and Tetracycline (97.3%), Tobramycin (89.3%) While they were sensitive to colistin and tigecycline. Partial agree with 24 who showed A. baumannii isolate absolute resistance (100%) to Ampicillin, Ceftriaxone, Ciprofloxacin, Cefotaxime, Gentamicin, Piperacillin, Ticarcillin, Trimethoprim-sulfamethoxazole. In comparison, it showed moderate resistance (50%) to Tobramycin and low resistance to Amikacin. COVID-19 patients admitted to ICU had bacterial coinfection with Gram-negative bacteria such as A. baumannii that resist all tested antibiotics except colistin 25 Virulence factors have the potential to alter antibiotic susceptibility profiles and confer drug resistance in A. baumannii through a variety of mechanisms. Some of these mechanisms include the regulation of antibiotic transportation through bacterial membranes, the modification of the antibiotic target site, and enzymatic modifications that result in antibiotic neutralization 26. The results showed Figure (4 ) high XDR (77.8%), MDR( 19.4%), and PDR( 2.8%), according to the categorized classification of antibiotic resistance. These results agree with a local study that showed resistance of A. baumannii clinical isolates to the tested antibiotics from 124 isolates with high XDR(75%), MDR (23.4%), and PDR (1.6%)27.
28 Discovered that 124 (100%) A. baumannii isolates were multidrug-resistant (MDR), 96 % of isolates were extensively drug-resistant (XDR). None of the isolates pan drug. 29 obtained 337 patients with A. baumannii ventilator-associated pneumonia (VAP); the proportion of MDR, XDR, and PDR were 21.4%, 65.3%, and 3.6%, respectively.
Acinetobacter baumannii, a multidrug-resistant strain of non-fermentative Gram-negative bacilli, has become increasingly common in hospital-acquired illnesses in recent years as a direct result of antibiotic overuse and the subsequent development of resistance to antibiotics (nosocomial infections). It would appear that assessing the drug susceptibility of resistant isolates is necessary to halt the spread of bacteria resistant to many drugs 30,31. The biofilm formation causes infections like ventilator-associated pneumonia and catheter-related infection, which can be highly resistant to antibiotic treatments and have become one of the most severe global problems due to the rapid spread of medical device-associated infections and antibiotic resistance 32. The Quantitative method( MTP) results showed that all the tested A. baumannii can form a biofilm at a rate of 100%. The study's results were identical to the recent local studies done by 33 obtained that (100%) of isolates formed Biofilm. It also found that all isolates formed Biofilm (100%).
Moreover, 35 reported that 97.1% of the clinical isolates form biofilms, while 36 reported that (76.8%) isolates were biofilm formers. In this research, we have observed no significant differences between the mean values of Biofilm after 48 hours among comparative groups with and without COVID-19 infection in the MTP method and the results obtained by the qualitative(CRA)method compatible with the study. He tested 16 isolates of A.baumannii, and only 13 could produce Biofilm. Furthermore, also agreement with the study by 38which found more A.baumannii isolates showed a strong slime layer on the Congo red agar 48(57.83%)and 39found 73.3% of isolates of A.baumannii were biofilm producers; however, 39who obtained out of 98 isolates, 7(7.4%) produced Biofilm including medium and robust and 91 (92.8) not produced. Several environmental parameters, including temperature, humidity, iron concentration, nutrients and quality of materials where biofilms are created light, and ambient acidic conditions, all affect the ratios of the biofilm development, which explains why proportions might vary from one research study to the next. 40 Antibiotics were unable to penetrate the Biofilm, and the structure of the Biofilm may prevent bactericidal concentrations from building up across the entirety of the Biofilm. The Biofilm's nutrition and oxygen distribution gradients, which result in unique metabolic states for individual cells, also promote antibiotic resistance and bacterial persistence41. However, there was a significant difference in adherence levels of bacteria using Congo red between the COVID-19 groups, with a more substantial proportion of solid adherence in the COVID-19 group compared to moderate commitment in the COVID-19-free group (87.2 %vs. 53.6 %). Patients with severe COVID-19 usually present the main risk factors observed to VAP caused by A. baumannii 42 Ventilator-Associated Pneumonia (VAP). COVID-19 infection can diagnosed by PCR43 and by CT44,45; the molecular technique was recommended to be applied in the different medical fields diagnosis of pathogenic bacteria, including Clostridium perfringens 46, Brucella melitensis 47, Proteus vulgaris 48,49, Staphylococcus aureus 50, Pseudomonas aeruginosa 51, and Toxoplasma spp 52,53, and some genetic disease ex: CML54, Adenocarcinoma 55,56.
CONCLUSIONS
There were no significant changes in mean biofilm values after 48 hours between groups with and without COVID-19 infection in the microtiter plate method. There was a significant difference in adherence levels of bacteria using Congo Red agar between COVID-19 groups, with a higher proportion of solid adherence in the COVID-19 group compared to moderate adherence in the COVID-19-free group patients.
Funding
No funds were received for this article.
Informed consent to publish
Not conflict
Acknowledgments: thank all your help in releasing this job.
REFERENCES
1. Wu T, Bian ZX, Abudu M, Adams D, Ko SG. Complementary and alternative medicine for respiratory tract infectious diseases: Prevention and treatments. Evidence-Based Complementary and Alternative Medicine. 2014 Jan 1;2014.
2. Al-Kaabi SK, Atherton A. Impact of noncommunicable diseases in Qatar. ClinicoEconomics and outcomes research: CEOR. 2015;7:377.
3. Manohar P, Loh B, Athira S, Nachimuthu R, Hua X, Welburn SC, Leptihn S. Secondary bacterial infections during pulmonary viral disease: phage therapeutics as alternatives to antibiotics. Frontiers in Microbiology. 2020 Jun 26;11:1434.
4. Doi Y, Murray GL, Peleg AY. Acinetobacter baumannii: evolution of antimicrobial resistance—treatment options. InSeminars in respiratory and critical care medicine 2015 Feb (Vol. 36, No. 01, pp. 085-098). Thieme Medical Publishers.
5. Lynch III JP, Zhanel GG, Clark NM. Infections due to Acinetobacter baumannii in the ICU: treatment options. InSeminars in Respiratory and Critical Care Medicine 2017 Jun (Vol. 38, No. 03, pp. 311-325). Thieme Medical Publishers.
6. Thummeepak, R., Kongthai, P., Leungtongkam, U. and Sitthisak, S., 2016. Distribution of virulence genes involved in biofilm formation in multidrug-resistant Acinetobacter baumannii clinical isolates. Int Microbiol, 19(2), pp.121-9.
7. Asfaw T. Biofilm Formation by Enterococcus Faecalis and Enterococcus Faecium. Int J Res Stud Biosci. 2019;7(4).
8. Runci F, Bonchi C, Frangipani E, Visaggio D, Visca P. Acinetobacter baumannii biofilm formation in human serum and disruption by the gallium. Antimicrobial agents and chemotherapy. 2017 Jan 1;61(1):e01563-16.
9. Antunes L, Visca P, Towner KJ. Acinetobacter baumannii: evolution of a global pathogen. Pathogens and disease. 2014 Aug 1;71(3):292-301.
10. Russo A, Bassetti M, Bellelli V, Bianchi L, Marincola Cattaneo F, Mazzocchetti S, Paciacconi E, Cottini F, Schiattarella A, Tufaro G, Sabetta F. Efficacy of a fosfomycin-containing regimen for treatment of severe pneumonia caused by multidrug-resistant Acinetobacter baumannii: a prospective, observational study. Infectious Diseases and Therapy. 2021 Mar;10(1):187-200.
11. Bassetti M, Righi E, Vena A, Graziano E, Russo A, Peghin M. Risk stratification and treatment of ICU-acquired pneumonia caused by multidrug-resistant/extensively drug-resistant/pan drug-resistant bacteria. Current opinion in critical care. 2018 Oct 1;24(5):385-93.
12. Russo A, Giuliano S, Ceccarelli G, Alessandri F, Giordano A, Brunetti G, Venditti M. Comparison of septic shock due to multidrug-resistant Acinetobacter baumannii or Klebsiella pneumoniae carbapenemase-producing K. pneumoniae in intensive care unit patients. Antimicrobial Agents and Chemotherapy. 2018 Jun 1;62(6):e02562-17.
13. Serafim RB, Póvoa P, Souza-Dantas V, Kalil AC, Salluh JI. Clinical course and outcomes of critically ill patients with COVID-19 infection: a systematic review. Clinical Microbiology and Infection. 2021 Jan 1;27(1):47-54.
14. Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. The Lancet. 2020 Mar 28;395(10229):1054-62.
15. Lima WG, Brito JC, da Cruz Nizer WS. Ventilator-associated pneumonia (VAP) caused by carbapenem-resistant Acinetobacter baumannii in patients with COVID-19: Two problems, one solution? Medical Hypotheses. 2020 Nov;144:110139.
16. Ripa M, Galli L, Poli A, Oltolini C, Spagnuolo V, Mastrangelo A, Muccini C, Monti G, De Luca G, Landoni G, Dagna L. Secondary infections in patients hospitalized with COVID-19: incidence and predictive factors. Clinical Microbiology and Infection. 2021 Mar 1;27(3):451-7.
17. Silva DL, Lima CM, Magalhaes VC, Baltazar LM, Peres NT, Caligiorne RB, Moura AS, Fereguetti T, Martins JC, Rabelo LF, Abrahão JS. Fungal and bacterial coinfections increase the mortality of severely ill COVID-19 patients. Journal of Hospital Infection. 2021 Jul 1;113:145-54.
18. Forbes BA, Sahm DF, Bailey WR, Weissfeld AS, Scott EG. Bailey & Scott's diagnostic microbiology. Mosby; 2007.
19. Drew WL, Barry AL, O'Toole R, Sherris JC. Reliability of the Kirby-Bauer disc diffusion method for detecting methicillin-resistant strains of Staphylococcus aureus. Applied Microbiology. 1972 Aug;24(2):240-7.
20. CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
21. Karimi S, Ghafourian S, Kalani MT, Jalilian FA, Hemati S, Sadeghifard N. Association between toxin-antitoxin systems and biofilm formation. Jundishapur Journal of Microbiology. 2015 Jan;8(1).
22. Badmasti F, Siadat SD, Bouzari S, Ajdary S, Shahcheraghi F. Molecular detection of genes related to biofilm formation in multidrug-resistant Acinetobacter baumannii isolated from clinical settings. Journal of medical microbiology. 2015 May 1;64(5):559-64.
23. Smail SB, AL-Otrachi KI. Phenotypic Characterization of Extended-Spectrum Beta-Lactamases and Metallo-Beta-Lactamase of Multi Drug Resistant Acinetobacter baumannii Causing Nosocomial Infections in Erbil City. Al-Mustansiriyah J Sci. 2020 Jan 15;30:51-6.
24. Wajeeh DN, Shareef AY, Shareef SY. Sensitivity Testing of Acinetobacter baumannii and Detection of some Resistance Genes Affiliate of blaoxa and adeABC. Rafidain Journal of Science. 2018 Dec 1;27(4):258-70.
25. Bazaid AS, Barnawi H, Qanash H, Alsaif G, Aldarhami A, Gattan H, Alharbi B, Alrashidi A, Al-Soud WA, Moussa S, Alfouzan F. Bacterial coinfection and antibiotic resistance profiles among hospitalized COVID-19 patients. Microorganisms. 2022 Feb 23;10(3):495.
26. Kyriakidis I, Vasileiou E, Pana ZD, Tragiannidis A. Acinetobacter baumannii antibiotic resistance mechanisms. Pathogens. 2021 Mar;10(3):373.
27. Ridha D, Ali M, Jassim K. Occurrence of Metallo-β-Lactamase Genes among Acinetobacter Baumannii Isolated from Different Clinical Samples. J. Pure Appl. Microbiol. 2019 Jun 1;13:1111-9.
28. Bagheri Josheghani S, Moniri R, Firoozeh F, Sehat M, Dasteh Goli Y. Susceptibility pattern and distribution of oxacillinases and blaPER-1 genes among multidrug-resistant Acinetobacter baumannii in a teaching hospital in Iran. Journal of Pathogens. 2015 Oct;2015.
29. Inchai J, Pothirat C, Bumroongkit C, Limsukon A, Khositsakulchai W, Liwsrisakun C. Prognostic factors associated with mortality of drug-resistant Acinetobacter baumannii ventilator-associated pneumonia. Journal of Intensive Care. 2015 Dec;3(1):1-8.
30. Royer S, de Campos PA, Araújo BF, Ferreira ML, Gonçalves IR, Batistão DW, Brígido RT, Cerdeira LT, Machado LG, de Brito CS, Gontijo-Filho PP. Molecular characterization and clonal dynamics of nosocomial bla OXA-23 producing XDR Acinetobacter baumannii. PLoS One. 2018 Jun 11;13(6):e0198643.
31. Mirzaei B, Bazgir ZN, Goli HR, Iranpour F, Mohammadi F, Babaei R. Prevalence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) phenotypes of Pseudomonas aeruginosa and Acinetobacter baumannii isolated in clinical samples from Northeast of Iran. BMC research notes. 2020 Dec;13(1):1-6.
32. Yang CH, Su PW, Moi SH, Chuang LY. Biofilm formation in Acinetobacter Baumannii: genotype-phenotype correlation. Molecules. 2019 Jan;24(10):1849.
33. Ahmad NH, Mohammad GA. Evaluation of Some Material to Inhibit Biofilm Formed by Acinetobacter baumannii Isolates. Tikrit Journal of Pure Science. 2019 Aug 4;24(4):19-24.
34. Al-Kadmy IM, Ali AN, Salman IM, Khazaal SS. Molecular characterization of Acinetobacter baumannii isolated from the Iraqi hospital environment. New microbes and new infections. 2018 Jan 1;21:51-7.
35. Al-Shamiri MM, Zhang S, Mi P, Liu Y, Xun M, Yang E, Ai L, Han L, Chen Y. Phenotypic and genotypic characteristics of Acinetobacter baumannii enrolled in the relationship among antibiotic resistance, biofilm formation and motility. Microbial pathogenesis. 2021 Jun 1;155:104922.
36. Castilho SR, Godoy CS, Guilarde AO, Cardoso JL, André MC, Junqueira-Kipnis AP, Kipnis A. Acinetobacter baumannii strains isolated from patients in intensive care units in Goiânia, Brazil: Molecular and drug susceptibility profiles. PLoS One. 2017 May 5;12(5):e0176790.
37. Al-Dulaimi AA, Al-Taai HR, Al-Bajlany SM. Virulence Factors of Acinetobacter baumannii isolated from different clinical specimens in Baquba. Diyala Journal For Pure Science. 2017;13(1-part 1).
38. AL-Mousawi HT, AL-Taee MI, AL-Hajjar QN. Evaluation of Biofilm Formation Capacity of Acinetobacter baumannii Isolated from Clinical Samples in Baghdad Hospitals using Phenotypic Methods. Iraqi journal of biotechnology. 2018;17(3). BibTeX
39. Avila-Novoa MG, Solís-Velázquez OA, Rangel-Lopez DE, González-Gómez JP, Guerrero-Medina PJ, Gutiérrez-Lomelí M. Biofilm formation and detection of fluoroquinolone-and carbapenem-resistant genes in multidrug-resistant Acinetobacter baumannii. Canadian Journal of Infectious Diseases and Medical Microbiology. 2019 Dec 20;2019.
40. Gedefie A, Demsis W, Ashagrie M, Kassa Y, Tesfaye M, Tilahun M, Bisetegn H, Sahle Z. Acinetobacter baumannii biofilm formation and its role in disease pathogenesis: A review. Infection and Drug Resistance. 2021;14:3711.
41. Uruén C, Chopo-Escuin G, Tommassen J, Mainar-Jaime RC, Arenas J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics. 2020;10 (1):3.
42. Karakuzu Z, Iscimen R, Akalin H, Girgin NK, Kahveci F, Sinirtas M. Prognostic risk factors in ventilator-associated pneumonia. Medical science monitor: international medical journal of experimental and clinical research. 2018;24:1321.
43. Rasoul LM, Nsaif MM, Al-Tameemi MT, Al-Rubaii BA. Estimation of primer efficiency in multiplex PCR for detecting SARS-Cov-2 variants. Bionatura, 2022, 7(3). http://dx.doi.org/10.21931/RB/2022.07.03.49.
44. Ahmed AW, RN, Naif MM, Yahya MT, Maulood KS, Alchalabi GB, Mohammed AG. Chest CT findings and experience in 100 COVID-19 patients in Mosul city, Iraq. Biomedicine. 2021;41(4):793-798.
45. Suhail HA, Abdulrahman DM, Ahmed AW. Fetal Biometry and Doppler Assessment of Pregnant Women with COVID-19. International Journal of Biomedicine, 2022, 12(4):554–559.
46. Hashim ST, Fakhry SS, Rasoul LM, Saleh TH, Alrubaii BA. Genotyping toxins of Clostridium perfringens strains of rabbit and other animal origins. In the Tropical Journal of Natural Product Research, this link is disabled. 2021;5(4):613-616.
47. abdulkaliq Awadh H, Hammed ZN, Hamzah SS, Saleh TH, AL-Rubaii BA. Molecular identification of intracellular survival related Brucella melitensis virulence factors. Biomedicine. 2022;42(4):761-5.
48. Abdul-Gani MN, Laftaah BA. Purification and characterization of chondroitinase ABC from Proteus vulgaris, an Iraqi clinically isolate. Current Science. 2017:2134-40.
49. kadhim AL-Imam MJ, AL-Rubaii BA. The influence of some amino acids, vitamins and anti-inflammatory drugs on the activity of chondroitinase produced by Proteus vulgaris caused urinary tract infection. Iraqi Journal of Science. 2016:2412-21.
50. Sabah Fakhry S, Noori Hammed Z, Abdullah Bakir W, Abdullah Laftaah ALRubaii B. Identification of methicillin-resistant strains of Staphylococcus aureus isolated from humans and food sources by Use of mecA 1 and mecA 2 genes in Pulsed-field gel electrophoresis (PFGE )technique. Revis Bionatura 2022; 7 (2) 44. http://dx.doi.org/10.21931/RB/2022.07.02.44.
51. Shehab ZH, AL-Rubaii BA. Effect of D-mannose on gene expression of neuraminidase produced from different clinical isolates of Pseudomonas aeruginosa. Baghdad Science Journal. 2019;16(2).
52. Abdulla L, Ismael MK, Salih TA, Malik SN, Al-Rubaii BA. Genotyping and evaluation of interleukin-10 and soluble HLA-G in abortion due to toxoplasmosis and HSV-2 infections. Annals of parasitology. 2022;68(2):385-90.
53. Jiad AL, Ismael MK, Muhsin SS, Al-Rubaii BA. ND2 Gene Sequencing of Subfertile Patients Recovered from COVID-19 in Association with Toxoplasmosis. Bionatura, 7(3), 45. http://dx.doi.org/10.21931/RB/2022.07.03.45.
54. Al-Musawi AH, Aziz HM, Khudair S, Saleh TH. Molecular characterization of HBB gene mutations in beta-thalassemia patients of Southern Iraq. Biomedicine. 2022 Nov 14;42(5):1040-1043 DOI. 10.21931/RB/2022.07.03.41.
55. Ali SM, Lafta BA, Al-Shammary AM, Salih HS. In vivo oncolytic activity of non-virulent Newcastle disease virus Iraqi strain against mouse mammary adenocarcinoma. AIP Conference Proceedings, 2021; 2372(1): 030010.
56. Ali SM, Laftah BA, Al-Shammary AM, Salih HS. Study the role of bacterial neuraminidase against adenocarcinoma cells in vivo. InAIP Conference Proceedings.2021;2372 (1): 030009.
Received: 28 May 2023/ Accepted: 15 July 2023 / Published:15 September 2023
Citation: Mutashar S S, Al-Mudallal N H A L, Ridha D J. Quantitative and Qualitative Determination of Biofilm Formation Pattern in Multidrug Resistance Acinetobacter baumannii in Correlation with COVID-19 and Respiratory Infected Patients. Revis Bionatura 2023;8 (3) 27. http://dx.doi.org/10.21931/RB/2023.08.03.27