2021.06.03.15
Files > Volume 6 > Vol 6 No 3 2021
INVESTIGATION / RESEARCH
Molecular characterization of netB and tpeL virulence factors and antimicrobial resistance genes of Clostridium perfringens isolated from herbs and spices.
Ashraf A. Abd El-Tawab1, Fatma I. El-Hofy1, Mohamed A. Abdelmonem2, Hend S. Youssef2*
Available from: http://dx.doi.org/10.21931/RB/2021.06.03.15
ABSTRACT
The present study aimed to determine some virulence-associated genes and antimicrobial multidrug resistance of Clostridium perfringens recovered from herbs and spices widely distributed in the Egyptian market. C. perfringens virulence and resistance factors were determined using PCR targeting the netB, tpeL, ermB, bla and tetK genes. Thirty three out of 392 samples (8.42%) from herbs and spices submitted to our laboratory for bacteriological screening were positive for presence C. perfringens. PCR results for the tpeL gene in isolated C. perfringens revealed 9 out of 33 (27.3 %) of isolates, while netB was not detected. The isolates were resistant to Clindamycin, Vancomycin, tetracycline, and erythromycin with inhibition zones of 6.28 ± 0.63, 8.78 ± 0.41, 9.63 ± 0.63, and 9.84 ± 0.66 mm, respectively. The genes mentioned above were selected to correspond to the ineffective antimicrobials; ermB for erythromycin, tetK for tetracycline, and bla for the remainder. PCR results for antibacterial resistant genes in isolated C. perfringens revealed their presence. From 33 isolates, bla gene was detected in 21 (63.4 %), tetK in 13 (39.4 %) and ermB in only one isolate (3.03 %). Sequencing analysis was done for the bla gene as an example for the detected genes as detected at the highest incidence (63.4%). No cross-relationship was detected upon comparing incidence data of both studied virulence genes and those of antimicrobial resistance. The present findings may explain the resistance of C. perfringens to the examined antibacterials and recommend avoiding the application of them to control the microbe. In addition, the authors recommend following strict hygienic procedures during the industry of herbs and spices to ensure their clearance from Clostridium perfringens before distributing the products as food additives into the markets.
Keywords: Clostridium perfringens, Herbs, Spices, Antibiotic Susceptibility Test, Resistance, Genes
INTRODUCTION
Among food additives, herbs and spices are considered as severe vectors for foodborne microorganisms, including C. perfringens 1; 2. Incidence of C. perfringens in herbs and spices has been checked worldwide, including India 3, Turkey 4, the United Kingdom 5, Italy 6, Lebanon 7, Saudi Arabia 8, and Egypt 9.
Abundant toxin production and multidrug resistance are considered as the two major problems caused by Clostridium perfringens. This microbe is Gram-positive eubacteria, a rod-shaped, spore-forming anaerobe widely distributed elsewhere in nature, including soil, surfaces, sewage, feces, foods, and food additives 10.
C. perfringens is a highly toxicogenic bacterium as it can produce various toxins (at least 17) that are considered to be its pathogenic virulence factors 11. Four among these toxins are commonly categorized as significant as they may cause lethality; these are α-, ß-, £-, and i-toxins. Each strain with its particular toxins is associated with a particular disease in humans and animals 12. For this, most studies are focused on looking at these toxins from bacterial isolates, including one carried out by our team (El-Tawab et al., 2021, under publishing). On the other hand, some other genes were considered minor ones because they are thought not to be pivotal for C. perfringens pathogenesis. Among these genes are found netB and tpeL toxins. However, recently, these genes were suggested to contribute to disease pathogenesis. For many years, α-toxin was thought to be the major virulence factor involved in necrotic enteritis, but a clostridial strain cloned for deactivation of α-toxin still able to produce lesions in broilers. This study led to the discovery of the pathogenicity of a new toxin, NetB 13. In addition, it was discovered that inoculation of broilers with strains positive for both netB and tpeL were associated with greater severity of gross lesions over strains with only netB 14, a fact that supports the idea of the pathogenicity of tpeL too.
Even though a relatively limited number of studies focused on investigating these two toxins and their driving genes, no study has looked at these two virulence factors in clostridial isolates from herbs and spices.
The second major problem of the studied microbe is its resistance to drugs. In a previous study, we have conducted the antibiotic susceptibility test and detected the resistance of clostridial isolates to Clindamycin, Vancomycin, tetracycline, and erythromycin. However, the underlying mechanism of this resistance was not investigated 9.
The present study aimed to investigate the netB and tpeL toxin/gene positivity of C. perfringens isolated from herbs and spices retailed all over the Egyptian markets and to determine the molecular mechanisms underlying the antimicrobial drug resistance by investigating the positivity to bla, tetK, and ermB genes. To fulfill this aim, PCR and sequencing, and phylogenetic analyses have been conducted on C. perfringens isolates from herbs and spices.
METHODS
Samples and isolates
The study was applied to 392 samples obtained from the top herbs and spices suppliers in Egypt. The samples have been screened for the incidence of C. perfringens. Thirty-three C. perfringens isolates were obtained from these samples following ISO 6887-2, ISO 6887-3, ISO 6887-4, or ISO 8261 9. The isolates were resistant to Clindamycin, Vancomycin, tetracycline, and erythromycin 9.
DNA extraction
DNA was extracted from the pure isolated colonies using the QIAamp DNA Mini kit purchased from Qiagen® (Hilden, Germany) following the manufacturer's instructions with some modifications. The kit provides silica-membrane-based nucleic acid purification from different types of samples. The spin-column procedure does not require mechanical homogenization; thus, the total hands-on preparation time is only 20 minutes. The DNA concentration was determined using NanoDrop ND-1000 Spectrophotometer from Thermo-scientific (Waltham, MA, USA) by reading absorbances at 260 and 280 nm. The extracted samples were stored at –20 °C until used as templates for PCR amplification.
PCR
The conventional PCR was applied to screen netB and tpeL toxin-encoding genes and those of resistance, namely, bla, tetK and ermB in the 33 isolates of C. perfringens, using Emerald Amp GT PCR Master Mix kit, Code No. RR310A was purchased from Takara Bio Inc.® (Shiga, Japan). The amplification process was performed according to the manufacturer's instructions, using specific primers from Midland® (TX, USA) and a thermal cycler from Biometra® (Jena, Germany). The thermal cycling conditions are briefly described in tables 1, 2, and 3.

Table 1. Oligonucleotide primers for the 5 targeted genes

Table 2. Preparation of 5 Clostridium genes uniplex PCR Master Mix
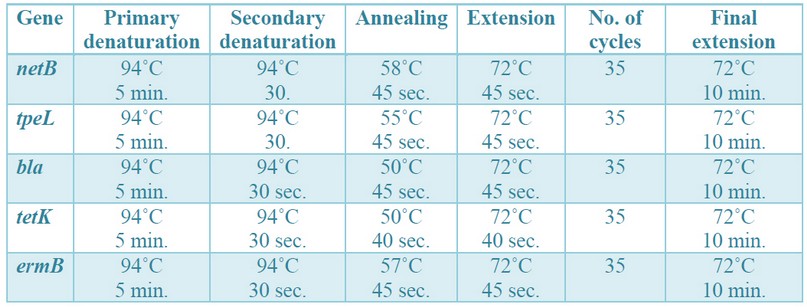
Table 3. Cycling conditions of the different primers for cPCR and RAPD
Agarose gel electrophoresis
Thirty μl of each PCR test product, negative and positive controls, and 100-bp DNA ladder (purchased from Fermentas®, Massachusetts, USA; cat. no. SM0243) were loaded to agarose gel 1.5 % and the process was conducted according to 20 following instructions of the manufacturer. The power supply was adjusted between 1~5 volts/cm of the tank length. The run was stopped after about 30 min, and then the gel was transferred to the UV cabinet. A gel documentation system photographed the gel, and the data was analyzed through computer software.
Sequencing reaction of bla gene
Uniplex PCR products of five C. perfringens isolates positive of bla gene were taken randomly and purified using the QIAquik PCR product purification protocol (Qiagen®, Hilden, Germany) provided by the manufacturer. The purified PCR products were sequenced in the forward and reverse directions on an Applied Biosystems 3130 automated DNA Sequencer (ABI, 3130, USA), using a ready reaction Bigdye Terminator V3.1 cycle sequencing kit, Cat. No. 4336817 (Perkin-Elmer / Applied Biosystems, Foster City, CA). The master mix using Big dye Terminator V3.1 cycle sequencing kit is described below (Table 4).
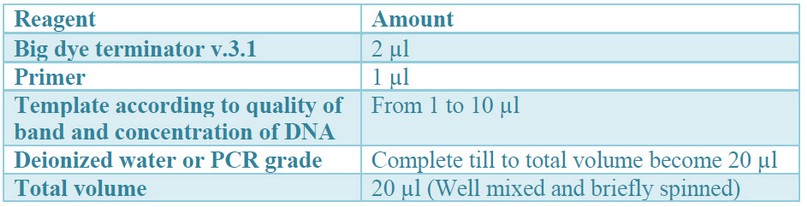
Table 4. Preparation of master mix using Bigdye Terminator V3.1 cycle sequencing kit
Phylogenetic analysis
The nucleotide sequences of bla gene were compared with the sequences available at public domains using BLAST (Basic Local Alignment Search Tool) server to establish sequence identity to GenBank accessions 21. A comparative analysis of sequences was performed using the CLUSTAL W multiple sequence alignment program, version 1.83 of MegAlign module of Lasergene DNAStar software Pairwise, designed by 22 and phylogenetic analyses done using maximum likelihood, neighbor-joining, and maximum parsimony in MEGA6 23.
GenBank submission
The sequences of the bla gene have been deposited in the GenBank database under the following accession numbers: HY1 MT891107; HY2 MT891108; HY3 MT891109; HY4 MT891110; HY5 MT891111.
Data management
The obtained results were statistically analyzed using EXCEL® software version 16. The number of positive samples for each gene against the total number of examined samples was calculated as a percentage from the total.
RESULTS AND DISCUSSION
C. perfringens was first isolated by William Welch and George Nuttall at a Hospital in Baltimore, USA, following a postmortem autopsy from a dead patient and was termed as Bacillus aerogenes capsulatus 24. The microbe can secrete various toxins of pore-forming nature by causing conformational change and barrel formation through the lipid bilayer of the affected host cells 25. Among those toxins, 4 types are considered as significant, which are alpha, beta, epsilon, and iota; and according to the ability of a clostridial strain to secrete one or more of such significant toxins, C. perfringens was subtyped into five types, A, B, C, D and E 26.
Identification and characterization of C. perfringens isolated from various sources have been made by many researchers. In the present work, 33 positive isolates for the studied bacteria were obtained from herbs and spices (33 isolates/392 samples) commonly distributed in the Egyptian market 9. Newer virulence factors, including net B and tpeL have been raised 13,27. Relatively to the major ones (α-, ß-, £-, and i-toxins), these two factors have limited the number of studies on some source materials, especially poultry affected with necrotic enteritis. Moreover, no information is available about them from herbs and spices; therefore, we have encouraged to look at netB and tpeL from this source.
Irrational antimicrobial use has increased the antimicrobial resistance among bacterial
pathogens, including C. perfringens. Moreover, an antimicrobial may also contribute positively or negatively to virulence factors of a bacterium 28.
In the present study, electrophoresis of the obtained multiplex PCR products confirmed the amplification of the target primer sequences for the positive controls of netB (560 bp) and tpeL (466 bp) gene fragments, while no bands were detected in the lanes of the negative controls. Bands corresponding to the tpeL fragments were detected in 27.3 % (9 out of 33) of the tested isolates, and no bands were detected for the netB fragments (Table 5 and Figures 1 & 2). The results may indicate the low prevalence of tpeL positive clostridial strains isolated from herbs and spices and the absence of those with netB genes. Although no similar studies were conducted on herbs and spices in Egypt, the absence of netB may be supported by the fact that this gene is only found in C. perfringens strains of poultry and exceptionally in one isolate recovered from a cow in the USA 29. Comparatively, the presence of tpeL in about one-third of the obtained C. perfringens colonies may be partially consistent with30, where was reported a 37 % of prevalence of clostridial strains positive for tpeL genes from ostriches. Low prevalence of both genes, netB, and tpeL in Alabama farms was also reported in 31. No studies have been conducted on herbs and spices regarding these genes to discuss. Generally, netB has a similar molecular size to beta-toxin, hence the name netB (necrotic enteritis toxin B-like), which has cytotoxic activity in chicken 13. While tpeL is a member of the large clostridial toxins that is still poorly understood and needs further investigations, it also has cytotoxicity 27.
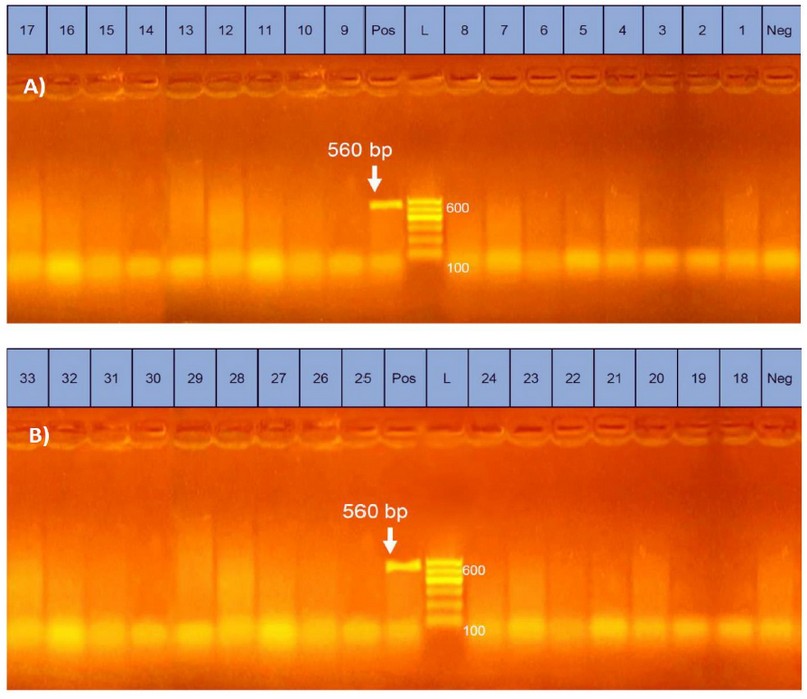
Figure 1. Uniplex PCR results of netB gene of C. perfringens isolated from herbs and spices in samples from 1~17 (A) and 18~33 (B); netB gene band was detected at 560 bp in control positive lane only.
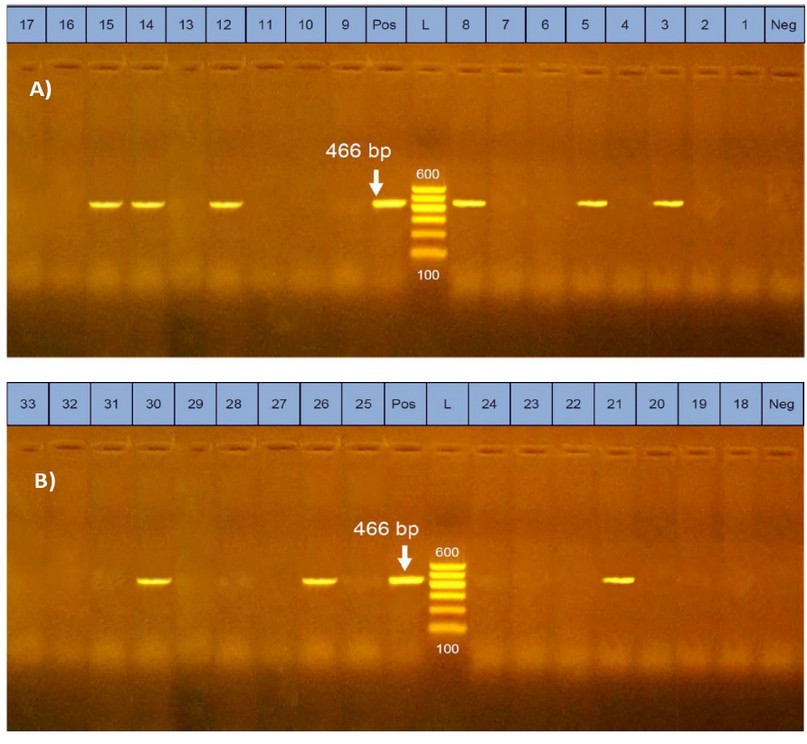
Figure 2. Uniplex PCR results of tpeL gene of C. perfringens isolated from herbs and spices in samples from 1~17 (A) and 18~33 (B); netB gene band was detected at 466 bp.
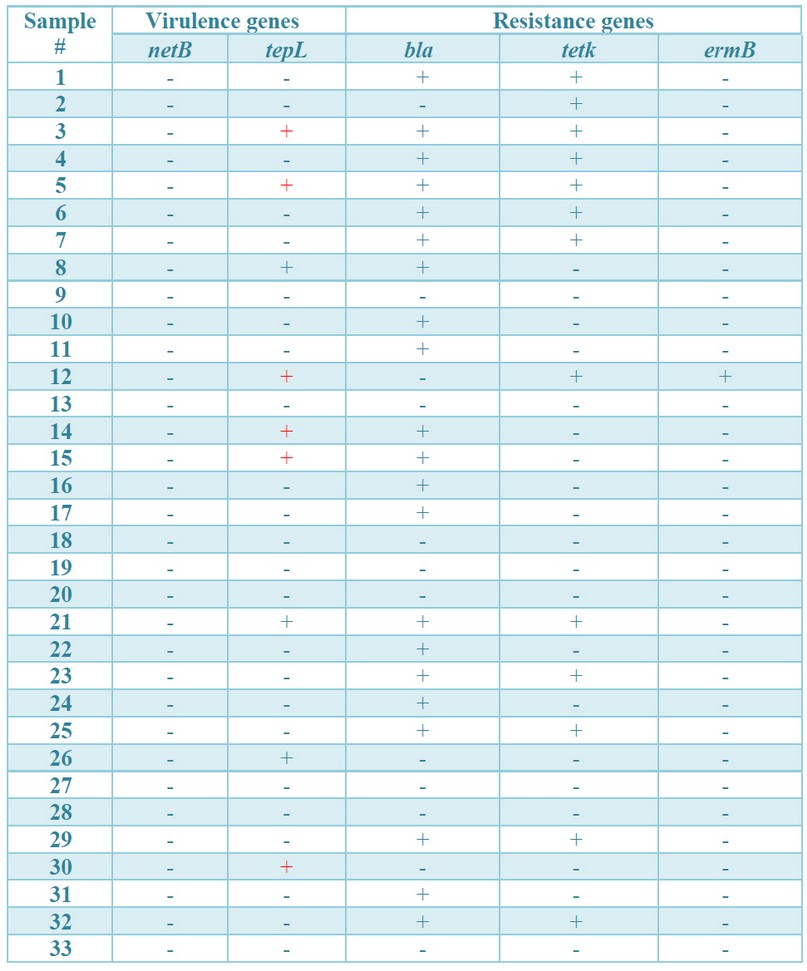
Table 5. Virulence (netB and tpeL) and multidrug resistance (bla, tetK and ermB) genes of C. perfringens isolated from herbs and spices
PCR data of the present study detected the presence of bla, tetK and ermB genes in C. perfringens isolates from herbs and spices. From 33 isolates, bla gene was detected in 21 (63.4 %), tetK in 13 (39.4 %) and ermB in only one isolate (3.03 %) (Table 5 and Figures 3, 4 & 5). This assay was aimed at exploring the genetic basis of our previous findings of antibiotic susceptibility test (AST) that found C. perfringens isolates resistant to Clindamycin, Vancomycin, tetracycline, and erythromycin with inhibition zones of 6.28 ± 0.63, 8.78 ± 0.41, 9.63 ± 0.63 and 9.84 ± 0.66 mm, respectively 9. The finding of bla may explain the resistance of C. perfringens to Clindamycin and Vancomycin based on its highest presence (about 64%), but the susceptibility to Penicillin-G (inhibition zone = 16.6 ± 1.16 mm) remains to be understood. The highest susceptibility of the microbe to Ampicillin-Salbactam (19.4 ± 0.98 mm) could be explained post-transcriptionally, where sulbactam inhibits beta-lactamase after its production from the bacterial cell. These findings may be in partial consistency with 32 who reported zero % resistance of C. perfringens to Penicillin, Cefoxitin, Meropenem, and piperacillin with 3.8 % of resistance to Clindamycin. However, our finding of resistance to vancomycin may be inconsistent with that of 33 34 who reported that C. perfringens to vancomycin, is low (0–5.6 %) because of the limited use of this antibiotic in farms. The finding of amplified bands of tetK gene fragments in C. perfringens isolated from herbs (39.4 %) may partially explain and parallel with the recorded resistance of isolates to tetracycline (8.8 ± 0.4 mm inhibition zone). In contrast, the finding of only 3 % of ermB-positive strains is not parallel with and cannot explain the resistance of isolates to erythromycin (9.8 ± 0.7 mm inhibition zone). This might refer to the presence of other mechanisms exhibited by the bacterium for resistance against erythromycin. The findings of resistance to Tetracycline and Erythromycin are consistent with 35 who reported resistance of C. perfringens isolates to these two antibiotics by 50.8 and 29.2 %, respectively. However, our finding may not agree with 36, who reported that the most probable mechanism of resistance against macrolides is modifying a target site by a methylase encoded by the ermB gene. The authors added that the ermB genotype exhibits the highest level of resistance against all macrolides, the statement does not match our findings as the ermB gene was detected in only one isolate among all the resistant isolates.
Taken together, it could be speculated that netB toxin does not correlate with the resistance of C. perfringens to the tested antibiotics because it was absent in all resistant isolates. However, tpeL toxin might contribute to the antimicrobial resistance as it was detected in about 40 % of the resistant isolates. Although this hypothesis needs more in-depth investigations, it could be in agreement with 35 who reported that the tpeL gene was more common among C. perfringens isolates susceptible to tetracycline. From our previous studies, it could be stated that the antimicrobial resistance profiles of C. perfringens varies significantly according to sources, countries, other bacterial factors rather than the resistance-driving genes.
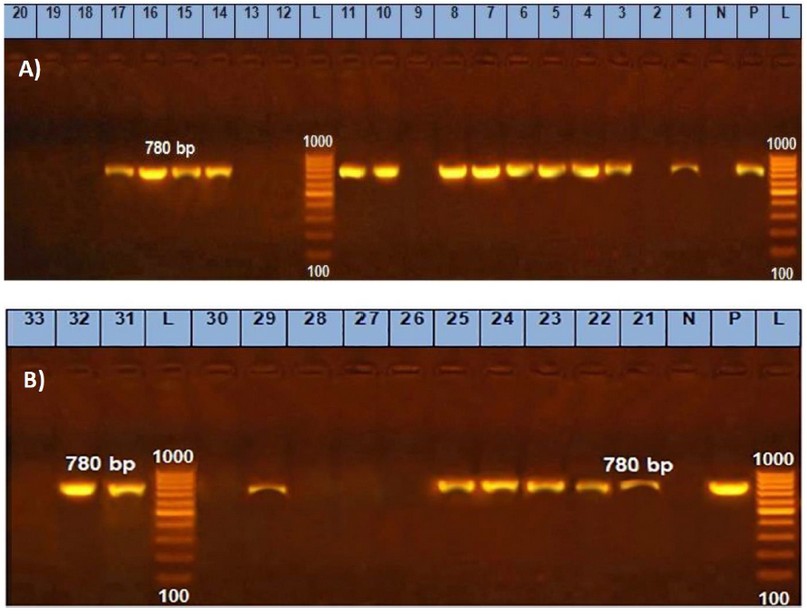
Figure 3. Uniplex PCR results of bla gene of C. perfringens isolated from herbs and spices in samples from 1~20 (A) and 21~33 (B); bla gene band was detected at 780 bp.
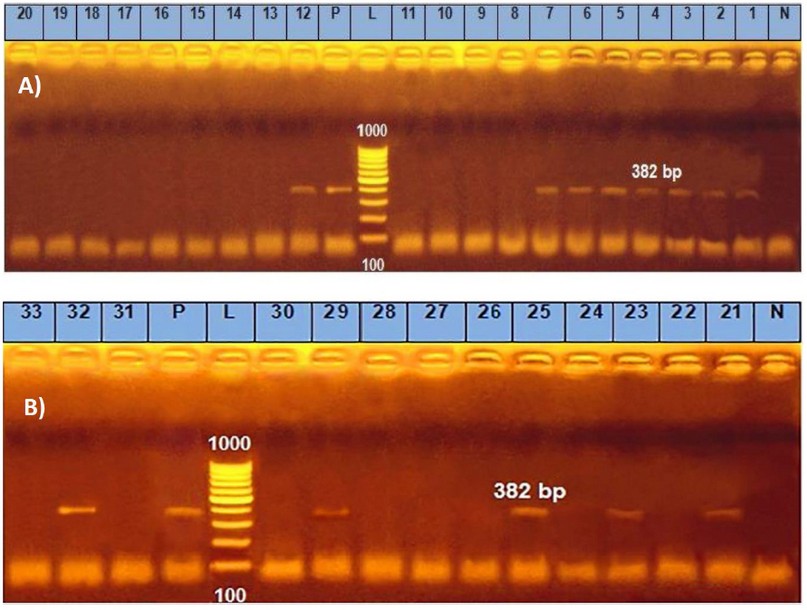
Figure 4. Uniplex PCR results of tetK gene of C. perfringens isolated from herbs and spices in samples from 1~20 (A) and 21~33 (B); bla gene band was detected at 382 bp.
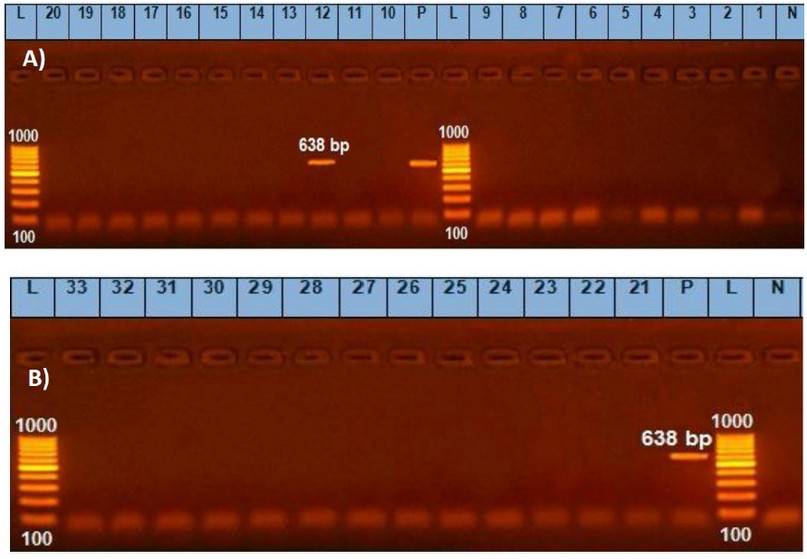
Figure 5. Uniplex PCR results of ermB gene of C. perfringens isolated from herbs and spices in samples from 1~20 (A) and 21~33 (B); bla gene band was detected at 638 bp.
Despite its apparent minimal contribution to antimicrobial resistance, yet, bla- gene was detected in 21 out of 33 isolates (63.4 %) of C. perfringens isolated from herbs and spices; therefore, its sequencing analysis becomes essential to map the epidemiology of C. perfringens infections caused by herbs and spices or food containing them. In the present study, sequence analysis was done for bla gene to detect its genetic diversity. The obtained sequences were analyzed using BLAST tool of GenBank. The BLAST result showed maximum identity ranging from 100% down to 97.3% with C. perfringens bla-gene. Our data (Figures 6 A & B) show the sequence alignment of the first 80 nucleotides from a total of 770 and the deduced amino acids of the first bla fragment, firstly isolated from herbs and spices. The overall sequence has high similarity and conservation with little divergences compared with those of bla-gene sequence of other global strains listed in GenBank.
Figure 7 depicts the phylogenetic analysis of bla-gene sequence, which shows a great degree of sequence conservation with slight divergence. The tree showed that the sequences of our local strain from herbs and spices have the same ancestors. There are no other sequencing studies conducted on isolates from herbs and spices to discuss our results with them.
Figure 6. Nucleotides (A) and amino acid (B) graphic view of bla sequencing analysis showing similarities represented by dots and diversities represented by letters. Gene sequences submitted by the authors are colored red.
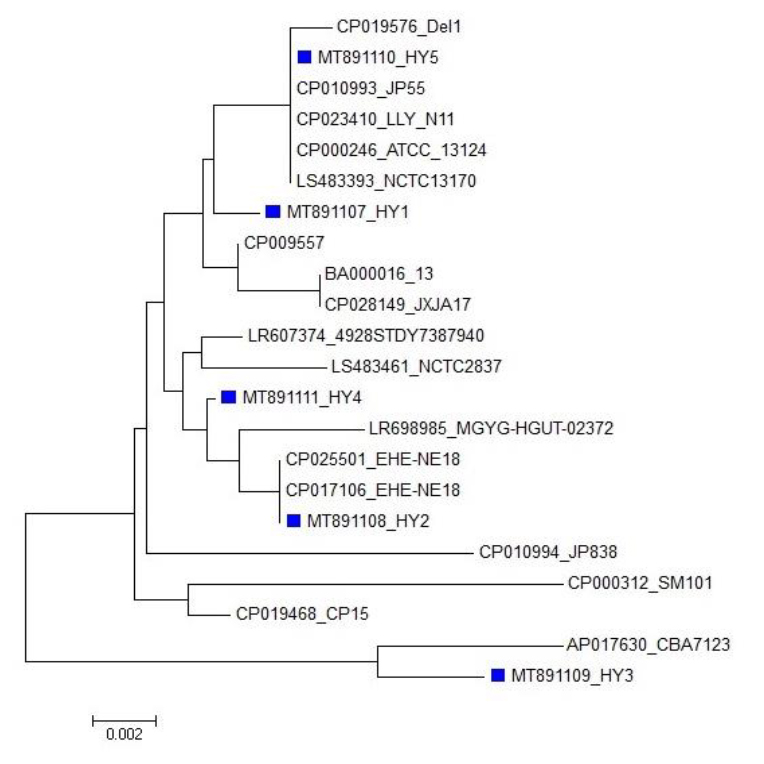
Figure 7. Phylogenetic analysis of nucleotide/amino acid sequences of bla coding gene of C. perfringens isolated from herbs and spices, the sequences of the present study are marked by blue squares among the sequences of GenBank.
CONCLUSION
From the data presented above, it could be concluded that strains of C. perfringens isolated from herbs and spices retailed in the Egyptian market have a higher occurrence of tpeL but not netB toxin coding genes. Resistance of C. perfringens to tetracycline is partially dependent on the presence of tetK gene. At the same time, sensitivity to Penicillin is evident despite the presence of bla-gene at a high rate among isolates that has a high similarity to the sequences submitted to GenBank among global isolates of epidemiological concern. These findings expand knowledge about C. perfringens isolated from herbs and spices as food additives, which provide the scientific basis for efficient prevention and intervention of C. perfringens-caused problems in man and animals.
Acknowledgments
Not applicable.
Declaration
Conflict of interest: There is no actual or potential conflict of interest concerning this article.
REFERENCES
1. Banerjee M, Sarkar PK. Growth and enterotoxin production by sporeforming bacterial pathogens from spices. Food Control 2004;15(6):491-496.
2. Aguilera MO, Stagnitta PV, Micalizzi B, de Guzmán AMS. Prevalence and characterization of Clostridium perfringens from spices in Argentina. Anaerobe 2005;11(6):327-334.
3. Banerjee M, Sarkar PK. Microbiological quality of some retail spices in India. Food Research International 2003;36(5):469-474.
4. Hampikyan H, Bingol EB, Colak H, Aydin A. The evaluation of microbiological profile of some spices used in Turkish meat industry. Journal of Food, Agriculture & Environment 2009;7(3&4):111-115.
5. Sagoo S, Little C, Greenwood M, et al. Assessment of the microbiological safety of dried spices and herbs from production and retail premises in the United Kingdom. Food microbiology 2009;26(1):39-43.
6. Vitullo M, Ripabelli G, Fanelli I, Tamburro M, Delfine S, Sammarco M. Microbiological and toxicological quality of dried herbs. Letters in applied microbiology 2011;52(6):573-580.
7. Debs-Louka E, El Zouki J, Dabboussi F. Assessment of the Microbiological Quality and Safety of Common Spices and Herbs Sold in Lebanon. J Food Nutr Disor 2 2013;4:2.
8. Hassan S, Altalhi A. Safety Assessment of Spices and Herbs Consumed In Saudi Arabia: Microbiological Quality and Toxin Production. Life Sci J 2013;10:2819-2827.
9. El-Tawab A, Abdallah M, Yusuf H. Incidence and antibiogram of Clostridium perfringens isolated from herbs and spices widely distributed in the Egyptian market. Benha Veterinary Medical Journal 2017;32(1):198-206.
10. Chen J, McClane BA. Characterization of Clostridium perfringens TpeL toxin gene carriage, production, cytotoxic contributions, and trypsin sensitivity. Infection and immunity 2015;83(6):2369-2381.
11. Bokori‐Brown M, Savva CG, da Costa SPF, Naylor CE, Basak AK, Titball RW. Molecular basis of toxicity of Clostridium perfringens epsilon toxin. The FEBS journal 2011;278(23):4589-4601.
12. Van Immerseel F, Rood JI, Moore RJ, Titball RW. Rethinking our understanding of the pathogenesis of necrotic enteritis in chickens. Trends in microbiology 2009;17(1):32-36.
13. Keyburn AL, Boyce JD, Vaz P, et al. NetB, a new toxin that is associated with avian necrotic enteritis caused by Clostridium perfringens. PLoS pathog 2008;4(2):e26.
14. Coursodon C, Glock R, Moore K, Cooper K, Songer J. TpeL-producing strains of Clostridium perfringens type A are highly virulent for broiler chicks. Anaerobe 2012;18(1):117-121.
15. Datta S, Rakha N, Narang G, Arora D, Mahajan N. Prevalence of α, β and netb toxin producing strains of Clostridium perfringens in broiler chickens in Haryana. Haryana Vet 2014;53(1):39-42.
16. Bailey MA, Macklin KS, Krehling JT. Use of a Multiplex PCR for the Detection of Toxin-Encoding Genes netB and tpeL in Strains of Clostridium perfringens. International Scholarly Research Notices 2013;2013.
17. Catalán A, Espoz M, Cortés W, Sagua H, González J, Araya J. Tetracycline and penicillin resistant Clostridium perfringens isolated from the fangs and venom glands of Loxosceles laeta: its implications in loxoscelism treatment. Toxicon 2010;56(6):890-896.
18. Gholamiandehkordi A, Eeckhaut V, Lanckriet A, et al. Antimicrobial resistance in Clostridium perfringens isolates from broilers in Belgium. Veterinary research communications 2009;33(8):1031-1037.
19. Soge O, Tivoli L, Meschke J, Roberts M. A conjugative macrolide resistance gene, mef (A), in environmental Clostridium perfringens carrying multiple macrolide and/or tetracycline resistance genes. Journal of applied microbiology 2009;106(1):34-40.
20. Sambrook J, Fritsch E, Maniatis T. Gel electrophoresis of DNA and pulsed-field agarose. Molecular cloning: a laboratory manual 1989;1:441-542.
21. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of molecular biology 1990;215(3):403-410.
22. Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic acids research 1994;22(22):4673-4680.
23. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular biology and evolution 2013;30(12):2725-2729.
24. Welsh W, Nuttall G. A Gas Producing Bacillus (Bacillus aerogenes capsulatus, nov, spec.) Capable of Rapid Development in the Blood Vessels after Death. Johns Hopkins Hospital Bulletin 1892;3:81-91.
25. Popoff MR. Clostridial pore-forming toxins: powerful virulence factors. Anaerobe 2014;30:220-238.
26. Songer JG. Clostridial enteric diseases of domestic animals. Clinical microbiology reviews 1996;9(2):216.
27. Amimoto K, Noro T, Oishi E, Shimizu M. A novel toxin homologous to large clostridial cytotoxins found in culture supernatant of Clostridium perfringens type C. Microbiology 2007;153(4):1198-1206.
28. Beceiro A, Tomás M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clinical microbiology reviews 2013;26(2):185-230.
29. Martin TG, Smyth JA. Prevalence of netB among some clinical isolates of Clostridium perfringens from animals in the United States. Veterinary microbiology 2009;136(1-2):202-205.
30. Mirzazadehghassab A, Razmyar J, Kalidari GA, Tolooe A. Prevalence of netb and Tpel Genes among Clostridium perfringens Isolates Obtained from Healthy and Diseased Ostriches (Struthio camelus). the 12th biennial Congress of the Anaerobe Society of the Americas2014.
31. Bailey M, Macklin K, Krehling J. Low prevalence of netB and tpeL in historical Clostridium perfringens isolates from broiler farms in Alabama. Avian diseases 2015;59(1):46-51.
32. Marchand-Austin A, Rawte P, Toye B, Jamieson FB, Farrell DJ, Patel SN. Antimicrobial susceptibility of clinical isolates of anaerobic bacteria in Ontario, 2010–2011. Anaerobe 2014;28:120-125.
33. Yanagihara K, Akamatsu N, Matsuda J, Kaku N, Katsumata K, Kosai K. Susceptibility of Clostridium species isolated in Japan to fidaxomicin and its major metabolite OP-1118. Journal of infection and chemotherapy 2018;24(6):492-495.
34. Li J, Zhou Y, Yang D, et al. prevalence and antimicrobial susceptibility of Clostridium perfringens in chickens and pigs from Beijing and Shanxi, China. Veterinary Microbiology 2021;252:108932.
35. Wei B, Cha S-Y, Zhang J-F, et al. Antimicrobial Susceptibility and Association with Toxin Determinants in Clostridium perfringens Isolates from Chickens. Microorganisms 2020;8(11):1825.
36. Roberts MC, Sutcliffe J, Courvalin P, Jensen LB, Rood J, Seppala H. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrobial agents and chemotherapy 1999;43(12):2823-2830.
Received: 11 March 2021
Accepted: 10 July 2021
Ashraf A. Abd El-Tawab1, Fatma I. El-Hofy1, Mohamed A. Abdelmonem2, Hend S. Youssef2*
1Department of Microbiology, Faculty of Veterinary Medicine, Benha University, 13736 Moshtohor, EGYPT;
2Department of Microbiology, Central Lab of Residue Analysis of Pesticides & Heavy Metals in Food, Agricultural Research Center, Ministry of Agriculture, 12311 Giza, Egypt.
*Corresponding author:
Hend Youssef,
Department of Microbiology,
Central Lab of Residue Analysis of Pesticides & Heavy Metals in Food,
Agricultural Research Center, Ministry of Agriculture, 12311 Giza, Egypt.
Email: [email protected]
Contact No.: +202 37611355
