2022.07.02.24
Files > Volume 7 > Vol 7 No 2 2022
1Department of Biology/College of Science/University of Mosul. Iraq
e-mail: [email protected]
2Department of Biology/College of Education/University of Mosul
* Correspondence: [email protected],
Available from: http://dx.doi.org/10.21931/RB/2022.07.02.24
ABSTRACT
This study (40) locally isolated the genus Streptomyces from soil samples collected from different regions of Iraq ( Nineveh , Erbil , Duhok ) and evaluated their antagonistic. The isolates were found to have bioactivity against gram-positive and negative bacteria and fungus. Streptomyces were isolated on (S.G. medium), and morphological similarities and the 16 srRNA sequencing were used to characterize them. . The results of a polymerase chain reaction (PCR) with eight strands of DNA gene picked from local bacteria isolates in a volume range of (900–1000) base pairs. The nitrogenic base sequence determined the polymerase chain reaction products of DNA samples selected from 6 local isolates. These strands preserved the employed DNA ladder volume. According to DNA Blast NCBI data, the species are Streptomyces atrovirens, Streptomyces SP.S. coeuleroubidus, and Streptomyces bellus.
Keywords. Streptomyces, Molecular, Pathogenic Microbes
INTRODUCTION
Among the microbe, actinomycetes are one of the critical sources for the production of antibiotics. About two–Streptomyces alone1 produce thirds of the antibiotics. Streptomyces are filamentous bacteria gram a positive with a high (G+C) content (70%) and found in all environmental2.
The genome code includes more than 20 genetic groups for secondary metabolites with high medicinal potential, such as antibiotics, which help fight against microbes3.
Many studies have found that Streptomyces has a considerable potential for producing secondary products such as antibiotics, growth factors, and pesticides4. Streptomyces can be distinguished from other actinobacteria by their 16 srDNA analysis and DNA - DNA hybridization5.
MATERIAL AND METHODS
Collecting of samples
(20) Soil samples were gathered from various farms in Nineveh at depths ranging from 5 to 10 cm below the earth's surface. Calcium carbonate (CaCO3) treated the samples at a 1:10 ratio. They were then dried for 4 days at (40–45) degrees centigrade. Samples are collected in plastic bags, closed tightly, and then transferred to a Freezing container until needed6.
Isolation of Streptomyces
(1) gram of dirt was well mixed in tubes with 10 ml of distilled water; then, a series of dilutions were performed. produced until the desired concentration was reached.10-6, 1 ml was taken from the last dilutions (10-5 - 10-6) and placed on culture medium Starch – casein medium (S.G.)
This was done three times for each sample (chilled to 45 degrees centigrade). The inoculum was then disseminated evenly using a sterile glass spreader and incubated for 7 days at 28 C0. Plates with 10–35 colonies were chosen, and the number of solitary colonies was re-cultured in the same medium to generate pure culture7,8.
The media
Starch – casein medium
It was made by using the following ingredients: (18) gm Agar (10) gm starch, (0.3) gm casein, (2) gm KNO3, (2) gm NaCl, (0.02) gm CaCO3, (2) gm KH2 PO4, (0.05) gm MgSO4 7H2 O (0.01) gram FeSO4. 7H2O, in (1) liter distilled water with a pH of (7.2). After that, the Autoclave apparatus was used to sterilize them9.
Nutrient agar medium
By melting 23 grams of the medium in (1) liter of distilled water with a pH of (7.2) and sterilizing everything in the autoclave, this medium was created according to (oxoid) the supply firm's recommendations
Starch mineral salts media
The following ingredients were used to make it: (10) gm Starch, (2) gm (NH4)2 SO4, and (2) gm CaCO3. (1) gram of K2 HPO4, (1) gram NaCl, (20) gm Agar in distilled water with a pH of (7.0), and they were all autoclave sterilized10.
2.3.4 Glycerol asparagine agar medium
Prepare this medium by dissolving: ( 1 ) gm asparagine, (10 ) gm glycerol, ( 1 ) gm K2HPO4, ( 20 ) gm agar, (1) ml of trace salt solution, ( 0.64 ) gm CuSO4. 5H2O. (0.11 ) gm FeSO4 . 7H2O, ( 0.79 ) gm MnCl2. 4H2O, ( 0.15) gm ZnSO4. 7H2O, The materials were dissolved in (1) liter of distilled water at pH (7.4) and sterilized in an autoclave11.
Antibiotics Production Medium
The medium for enhanced antibiotic synthesis was created by mixing (0.8) gm NaCl, (1) gm NH4Cl, (0.1) gm K2HPO4, (0.2) gm MgSO4 7H2O, (0.1) gm CaCl2, (10) gm glucose, and (3) gm yeast extract. The materials were dissolved in (1) liter of distilled water at pH (7.3) and sterilized in an autoclave12.
Isolation and characterization of Streptomyces
The Gram staining, colony morphology, and color of colonies, the starch mineral agar media, and using the slide culture technique been noticed the shape of the aerial and substrate mycelium, as well as the spore arrangement were used to characterize Streptomyces12.
Assay the antagonistic activity of isolates in this experiment
Staphloccocus aureus Proteus vulgris, E. coli , Klebsilla , Pseudomonas aeruginosa( and pathogenic fungi (Candida albicans, Rhizoctonia solani, Fusarium solani, Alternaria alternata .
Antifungal activity against phytopathogenic fungi
To evaluate the effect of Streptomyces isolates on the growth mycelium of plant pathogenic fungi, the Streptomyces isolates were transferred to the middle of a petri dish, distributed vertically in the petri dish by a sterile loop and incubated at 28-+1 0C for ( 5 ) days, after the incubation period two discs from the eight-day-old fungal culture were transferred to half dishes ( 1.5 ) cm away from the growth line of the Streptomyces isolates. The control treatment contains fungi discs only, which are incubated 28-+ 1 for 5 days ( the incubation period depends on the arrival of the mycelium fungi to the edges of the dish13.
Antibacterial activity against bacteria pathogen
The bioactivity of the Streptomyces was studied against the microbial Agar diffusion method14. Asparagine glycerol agar plates were inoculated with Streptomyces isolates and incubated at 28-+ 1 0C FOR ( 7) days. After the isolates were grown, 8 ( mm ) discs were taken by a sterile cork borer from the developing colonies of Streptomyces and transferred to the nutrient agar medium inoculation with pathogen bacteria.
Diagnosis of selected samples of bacteria Streptomyces
The selected Streptomyces was diagnosed to the species level using the diagnostic tests.15,12,16,17
Preparation of inoculation
The elements in the inoculation medium are the same as those in the production media. They were cultivated in autoclave-sterilized 250 mL flasks with 50 mL each. They were inoculated by transferring a tip of Streptomyces cultured on Glycerol asparagine agar to the inoculum. The flask was then shaken in a shaker incubator for 3 days at 140 rounds per second at (28-+1) Celsius.
Cultural Condition
In a 250 mL flask, 50 mL of antibiotic manufacturing media was created. The flasks were tightly sealed, and sterilization in the autoclave was conducted. They were all allowed to cool before injecting inoculum prepared from the 3-day-old chosen isolation at a 2:1 (v/v) ratio. The flaks were kept in an incubator vibrator for 7 days at (28+- 1) degrees Celsius and 240 round minutes.
Purification and acquisition of DNA from Streptomyces
The DNA from the Streptomyces samples was extracted using Geneaid's analysis kit.
PCR reactions: Genomic DNA Extraction
The T.E. (the solution was used to optimize the DNA concentration in all of the samples used in this experiment, and it was successful in obtaining the desired concentration for the PCR reaction to continue ( 50 nanograms per microliter ).
In a (0.2 ml) Eppendorf tube provided by the British business, the DNA sample was mixed with the specific primer for each gene to make the master reaction for each PCR reaction ( bio labs ). Using distilled water, the reaction volume was set to 20 microliters, and the reaction was placed in a microfuge for 3–5 seconds. The tubes were then placed within a heat cycler, which used a customized program for each reaction to achieve the polymer reactions. The samples were then electrophoresed using a bio labs ladder in a well of a 2 percent agarose gel for 60–70 minutes, and the gel was filmed using Gel documentation.
Streptomyces molecular diagnostics based on 16srRNA amplification.
To identify the amplification area, Add 4 microliters (100 nanograms) of DNA template and 1 microliter (10 picomols) of each gene primer were added to the final mix
Primer Forward: AAGCCC TGG AAACGGGGT
Primer Reverse : CGTGTGCAGCCC AAGACA
After that, the reaction tubes were placed in the thermal cycler, and the polymers reaction was carried out according to the program listed in the table. (1).
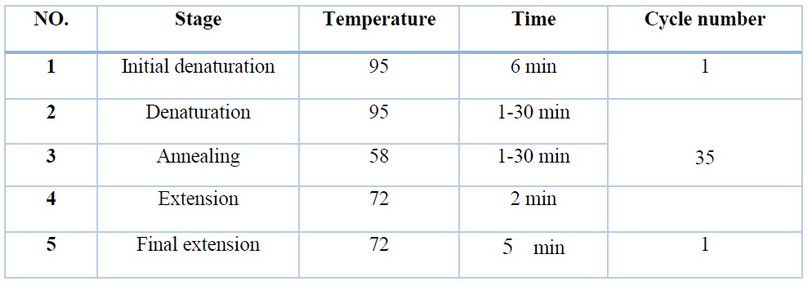
Table 1. Shows the PCR reaction inside the thermocycler device
DNA Sequencing analysis
DNA sequencing is the most frequently and commonly utilized technique for finding mutations and SNP variants in DNA samples. The sequence of amplified portions of DNA is used to determine, and PCR reactions usually determine research mutations. DNA sequencing results have gotten more precise in determining mutations in recent years18,19. If the PCR reaction produces many strands, they are purified, and the desired section of the DNA is recovered from the gel; however, if the reaction produces just one strand, it will be the predominant strand and will be used to identify the sequence20.
Using DNA sequencing to determine the nucleotides of the amplifier section.
The genetic sequences of the samples were determined using the PCR technique outlined previously. Used in the study, and the primers were analyzed with the Hitachi 3130 Genetic Analyzer device. The results were analyzed using the BLAST tool, which compared the genetic sequences to those in the National Center for Biotechnology Information) NCBI( database.
Diagnosis
The Streptomyces samples were identified using the slide culturing technique, one of the best techniques (on a genus level) to display substrate and hyphae, which are the differentiating traits that identify thread-like bacteria16.
The substrate hyphae are spore-free, well-branched, and unsegmented. While the aerial hyphae appeared to be a darker, thicker, and less branched thread than the substrate mycelium, the aerial hyphae have a sporophore containing chain of spores, which can be erect (rectus), spiral (spiral), or Crict with waves (rectus – flexible) depending on how these spores are arranged21. figure (1) .

Figure 1. The shape of the substrate and aerial mycelium of Streptomyces SPP
When the isolates were cultivated on different media types, they revealed a variety of colors. They could not produce melanin and other colors, with gray colonies being the most common21.

Figure 2. The shape of colonies and pigment production of Streptomyces
Genomic DNA polymers reaction
Using the forward primer, the following DNA,s reaction was performed on pure DNA acquired from organisms gathered from samples depending to the Geneaid protocol:
(AAGCCCTGGAAACGGGGT) And the revers:(CGTGTG CAGCCCAAGACA). (Maleki et al.,2013) In Figure 3, it can be seen that strands of pure DNA from the samples are of the same length (1000–9000) base pairs generated from the Streptomyces DNA specified polymer process. It has been discovered that they have the same length, indicating that there are nitrogenous base sequences in DNA that are mutual
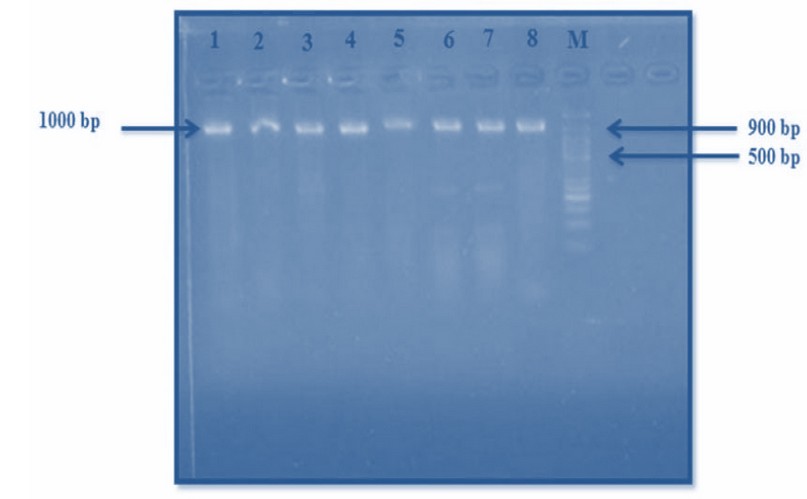
Figure 3. The specific polymerase reaction of Streptomyces spp. Primer 16s r RNA
These isolates' genomic DNA can link with the primer and continue the reaction, resulting in additional DNA strands of the same length. These results are similar to what was found22 When utilizing the polymers reaction approach to identify local bacterial samples and acquire a positive result, the length of the DNA created was (1000) nitrogenous base pairs, and so on. When using the polymers reaction method to analyze a bacterial local sample identification, the length of the DNA obtained was 1000 nitrogenous base pairs and so on (µ) marker evaluating and assessing the sequence of nitrogenous bases obtained from particular PCR., from local samples.
The findings of the specific polymer process on isolated DNA from local samples were forwarded to the NCBI, where they were compared to sequences in the gene bank figure (4).

Figure 4. Shows an analysis of the nitrogen base sequence of the gene bank
Antifungal activity against phytopathogenic fungi
In this study of the ability of Streptomyces isolates to inhibit plant pathogenic fungi Table (2). The isolates Streptomyces satrovirnes showed inhibit against fungi ( Fusarium solani , Rhizoctonia solani , Alternaria ignifica , Candida albicans ) , while the isolates S. coeuleorubidus , S. sp , S. bellus , inhibited fungi C. Albicans , F. solani , and A. alternate respectively Figure(5).
The Streptomyces can be biologically controlled by resistance / or killing fungi and bacteria pathogens; they are often Bioactive before the pathogen thoroughly infects its host. Recently a focus has been placed on different methods of developing biocontrol strategies against soil-borne fungi23. For example, the seed of the Arabidopsis thaliana.
Before sowing, treatment with Streptomyces and Micomonospora SP was protected from infection by Erwinina carotovra and F . oxysporum .
It has been observed that Streptomyces have an antagonistic ability to stimulate the defense mechanisms in the plant24.
In this study25. Indicated a signification inhibition of mycelium growth in dual culture, using three strains of Streptomyces. S.P.ALP 07R, Streptomyces S.P. MRA 1W, against six types of plant pathogenic fungi Sclerotinia sclerotiorum , Fusarium oxysporm ,f. S.P. Lactuca . Rhizoctonio solani Thielvopsis basicola , Phytophthora S.P. Pythium ultimum . According to 26, one strain of Streptomyces was found among Streptomyces' one hundred seven isolates. In vitro and in vivo, S.P. SI RO3 reduced the growth of mycelium Pestalotipsis theae by a percentage percent 86.15 percent 93.85, respectively.

Table 2. The effect of Streptomyces isolates of phytopathogenic fungi (mm)
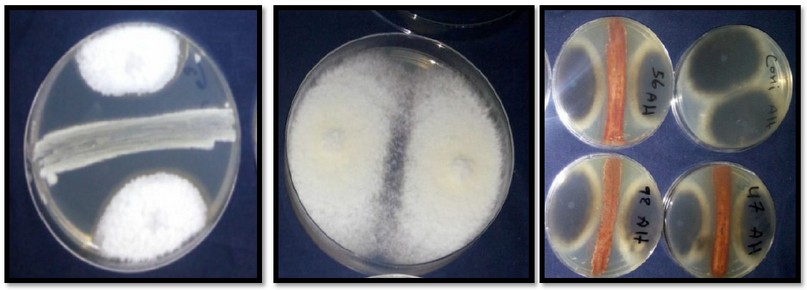
Figure 5. The effect of Streptomyces on phytopathogenic fungi
Antibacterial activity against bacteria pathogen
In this study, the biological activity of Streptomyces against some pathogenic bacteria was also evaluated. The Streptomyces Bellus give inhibition against all pathogenic bacteria used in this study, while the Streptomyces SP. and S.atrovirence were effective against all bacteria except Klebisla Penumonia Table (3). The researchers27. Showed that the inhibition extracted by acetate was effective against E SBL – producing E- coli . Numerous studies have indicated that the metabolites produced by actinomycetes showed high activity against Gram-negative, including E . coli28. The Streptomyces SMO1 gives a high inhibition against bacteria Staphylococcus aureus at a concentration ( 5 ml ) when grown on medium AIA at P.H. ( 7 ) for 7 days compared with Streptomycin ( 5 mg ) ampicillin ( 5 mg ) also indicated that among 8 isolates, one isolates S MO1 give inhibition against six bacteria strains except for K. pneumonia and S. typhimurium29. However, due to a double membrane barrier and brown transmembrane efflux, Gram-negative bacteria are more resistant to antimicrobial agents than Gram-positive bacteria28.
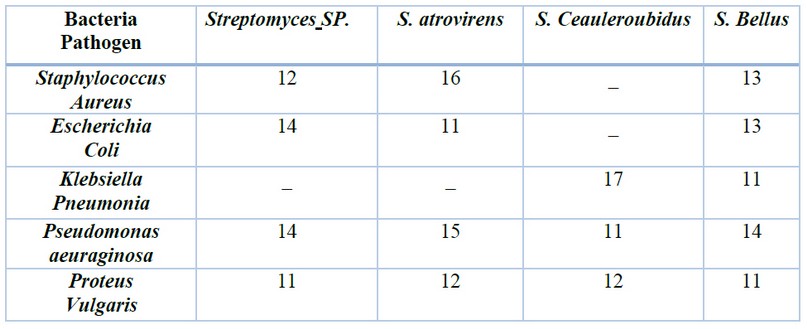
Table 3. The effectiveness of Streptomyces on bacteria pathogen inhibition zone ( mm )
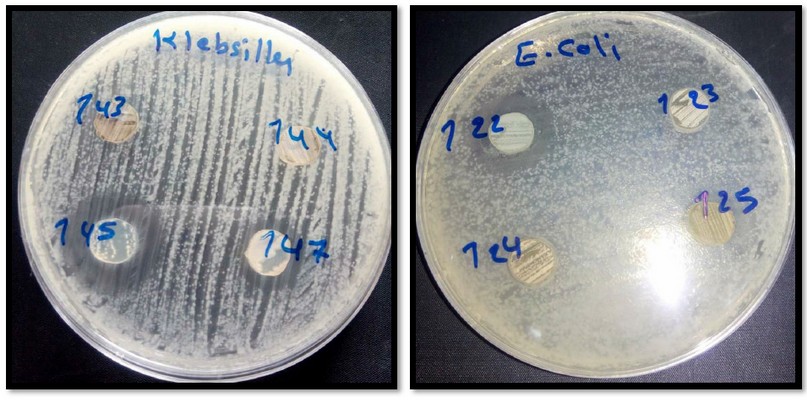
Figure 6. The effect of Streptomyces isolates on pathogenic bacteria.
CONCLUSION
Among the results obtained from both aspects, the molecular study proved its effectiveness and usefulness for plant breeders and the building block for the success of any plant breeding program and the shortening of time in determining the appropriate varieties to conduct crosses, improving the characteristics of the yield.
Funding: self-funding
Acknowledgments: In this section, we acknowledge any person who supports us to complete this project.
Conflicts of Interest: there is no conflict
REFERENCES
1. Liu ,X; Gu, k; Xia ,S ;Zhang ,D and Li ,y . Dolyemycin A and B , two novel cyclopiptide isolated from Streptomyces griseus Sub sp .griseus HYS31.J.of antibiotic, 2018: 71,838-845.
2.Bao , J; He, F; Fang ,L; Wang ,K; Song ,J ;Zhang ,J ; Li ,Q and Zhang , H . Cytotoxic antibiotic angucyclincs and actinomycin from Streptomyces sp. XZHG 99T .J. antibiotic ,2018: 71,1018-1024.
3.Hopwood, D. A. "Highlights of Streptomyces genetics".Heredity,2019: vol. 123, no. 1, pp. 23–32,.
4.Harir, M. Bendif, H. Bellahcene, M. Fortas, Z. and Pogni, R. "Streptomyces secondary metabolites" Basic Biology and Applications of Actinobacteria,2019: vol. 6, pp. 99–122.
5.Kumar, P. S. Raj, J. P. P. Duraipandiyan, V. and Ignacimuthu, S. "Antibacterial activity of some actinomycetes from Tamil Nadu, India," Asian Pacific Journal of Tropical Biomedicine,2012: vol. 2, no. 12, pp. 936–943.
6.Ceylan, O.; Okmen, G. and Ugur, A. Isolation of soil Streptomyces as source antibiotics active against antibiotic-resistant bacteria. EurAsian Journal of BioSciences,2008: 2(1), 73-82.
7.Al-Obaidi, S.I. Isolation of Different Local Species of the Genus Streptomyces and Study of their Antimicrobial Activity Identification by Biochemical Tests and Obtaining of Genetic Variation of Isolates by PCR RAPD Technique. Ph.D. Thesis, College of Education, University of Mosul. Mosul, Iraq.2012.
8.Ahmad, M.S.; El-Gendy, A.O.; Ahmed, R.R.; Hassan, H.M.; El-Kabbany, H.M. and Merdash, A.G. Exploring the antimicrobial and antitumor potentials of Streptomyces sp. AGM12-1 isolated from Egyptian soil. Frontiers in microbiology,2017: 8, 438.
9.Kuster, E. and Williams, S.T. Selective media for the isolation of Streptomycetes. Nature,1964: 202: 928-929.
10. Wu, R.Y. and Chen, M.H. Identification of the Streptomyces strain KS3-5. Botanical Bulletin of Academia Sinica,1995: 36.
11. Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T. and Williams, S.T.Endospore-forming grampositive rods and cocci. Bergey's Manual of Determinative Bacteriology. William and Wilkins, Baltimore USA,1994: 559.
12. Williams, S.T.; Goodfellow, M.; Alderson, G.; Wellington, E.M.H.; Sneath, P.H.A. and Sackin, M.J. Numerical classification of Streptomyces and related genera. Microbiology,1983: 129(6): 1743-1813.
13. Soares, A. C. F. ; Sousa, C. D. S. ; Garrido, M. D. S. ; Perez, J. O. and Almmedia, N. S. D. Soil Streptomyces within vitro activity against the yam pathogen Curvularia eragrostides and Colletetrichum gloeosporioides. Braz J. Microbiol.2006:vol.37.
14. Egrov, N. S. Antibiotics. A scientific approach. Mir Publishers.1985: Moscow, Russia.
15. Oskay, M.; Tamer A.U. and Azeri, C. Antibacterial activity of some actinomycetes isolated from farming soils of turkey. Afr J Biotechnol;2004: 3(9): 441-446.
16. Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T. and Williams, S.T.Endospore-forming grampositive rods and cocci. Bergey's Manual of Determinative Bacteriology. William and Wilkins, Baltimore USA,1970: 559.
17. Pridham, T.G. and Gottlieb, D. The utilization of carbon compounds by some Actinomycetales as an aid for species determination. Journal of bacteriology,1948: 56(1): 107
18. Smith, C.; Gore, A.; Yan, W.; Abalde-Atristain, L.; Li, Z.; He, C.; Wang, Y.; Brodsky, R.A.; Zhang, K.; Cheng, L. and Ye, Z. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN based genome editing in human iPSCs. Cell stem cell,2014: 15(1), 12-13
19. Cheng, L.; Hansen, N.F.; Zhao, L.; Du, Y.; Zou, C.; Donovan, F.X.; Chou, B.K.; Zhou, G.; Li, S. and Dowey, S.N. NISC Comparative Sequencing Program Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell,2012: 10(3): 337-344.
20. DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M. and McKenna, A. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics,2011: 43(5), 491.
21. Maleki, H.; Dehnad, A.; Hanifian, S. and Khani, S. Isolation and Molecular Identification of Streptomyces spp. With Antibacterial Activity From Northwest of Iran .Bio, 2013.
22. Carpenter, P.L. Growth and death of bacteria. Microbiology,1977: ed, 4, 217-229.
23. Samac, D.A.; Willert, A.M.; McBride, M.J.; Kinkel, L.L.Effects of antibiotic-producing Streptomyces on nodulation and leaf spot in Alfalfa. Appl. Soil. Ecol.2003: 22, 55–66.
24. Conn, V.M.; Walker, A.R.; Franco, C.M. Endophytic actinobacteria induce defense pathways in Arabidopsis thaliana. Mol. Plant Microbe Interact.2008: 21, 208–218, doi:10.1094/MPMI-21-2-0208.
25. Kunova ., A; Bonaldi.,M; Saracchi.,M; Pizzatt.,C; Chen., X; and Cortesi.,P. Selection of Streptomyces against soil borne fungal pathogenic by a standardized dual culture assay and evalution of their effects on seed germination and plant growth . BMC. Microbiology .2016: 16:272.
26. Marimuthu.,S; Karthic.,C; Mostafa .,A; AL-Enazi.,N-M ; Abdel-Raouf.,N and Sholkamy .,E-N. Antifungal activity of Streptomyces sp. SLRO3against tea fungal plant pathogen Pestalotiopsis theae .J. King Saud Uni –Sci.2020: 32.3258-3264.
27. Khadayat.,K, ;Sherpa., D-D; Malla., K-P; Shrestha.,S; Rana.,N; Marasini.,B-P; Khanal., S; Rayamajhee.,B; j Bhattarai.,B -R and Parajuli.,N. Molecular Identification and Antimicrobial Potential of Streptomyces Species from Nepalese Soil.2020: Int.J. Micro 8 page.
28. Zgurskaya ,. H. I and Nikaido, H. "Multidrug resistance mechanisms: drug efflux across two membranes," Molecular Microbiology,2000: vol. 37, no. 2, pp. 219–225.
29. Maiti,. P-k ; Das., S; Sahoo., P. and Mandal .,S. Streptomyces sp. SM01 isolated from Indian soil produces a novel antibiotic picolinamycin efective against multi drug-resistant bacterial strains, 2020: 10:10092. https://doi.org/10.1038/s41598-020-66984-w 1
Received: 7 December 2021 / Accepted: 25 January 2022 / Published:15 May 2022
Citation: Shaymaa khaleel alhialy, Arwa Shawkat Thanoon, Molecular Diagnosis of Streptomyces Genus and Bioactive Potential Against Pathogenic Microbes. Revis Bionatura 2022;7(2) 24. http://dx.doi.org/10.21931/RB/2022.07.02.24
