2022.07.03.25
Files > Volume 7 > Vol 7 No 3 2022
Submission and Phylogenetical of Local Isolated Trichophyton interdigitale of Iraqi Patients in NCBI.
College of Education for Pure Science Ibn-Al-Haitham / University of Baghdad / Baghdad -Iraq
Corresponding author: [email protected],
Available from: http://dx.doi.org/10.21931/RB/2022.07.03.25
ABSTRACT
Dermatophytes are species with slight genetic variation, and are yet several uncertainties about the differences among species. This study aims to isolate and diagnose the Trichophyton interdigitale by molecular technique and to reveal the phylogenetic distance and similarity of the Iraqi isolates to other isolates from the globe, in addition, to submit the obtained sequences to the NCBI database. This study included 86 with multiple lesions on different parts of the body. The results showed different variations within the ITS gene between the isolates. It was concluded that Trichophyton interdigitale in Iraqi isolates had two types of substitution variations (Transition and Transversion) different than global isolates. Moreover, it appeared according to the phylogenetic tree, the similarity was 97% with isolates from the Czech Republic and Japan, whereas 98% with the isolates from France and Germany.
Keywords. Dermatophytes, genetic variations, ITS gene, multiple lessons
INTRODUCTION
The keratinophilic group is categorized by several genera such as Trichophyton, Microsporum, Epidermophyton, Nannizzia, Paraphyton, Lophophyton, and Arthroderma; additionally, one of the well-known fungi which belong to the genera are dermatophytes. Those fungi can infect both humans and animals and can cause severe conditions 1
Dermatophytes are fungi that can cause dermatophytosis as they can produce protease and keratinase, which degrade keratin and thus lead to invasion and infection of different body sites. Genetic factors have a central role in the pathogenesis of dermatophytosis 2. One of the examples of anthropophilic is Trichophyton interdigitale, which causes dermatophytosis. Keratomycosis, also known as fungal keratitis, is a cornea infection. If this infection has been misdiagnosed or has been delayed to diagnosis can lead to loss of vision 3. Several studies have been done to know the incidence distribution in the different geographical areas of the world, including Iraq, specifically, Baghdad governarate, by Minnat and Jinan4, Jordan 5, Kuwait and Saudi Arabia mentioned in previous study 6.
The most well-known technique to diagnose and differentiate between species and sub-species is PCR-based assays, considered a quick method 7. Following PCR, sequencing is an effective method and informative for diagnosing fungal infections. Classification and taxonomy can be further improved by phylogenetic analysis of internal transcribed spacer (ITS) gene 8. Therefore, the current research aimed to isolate Trichophyton interdigitale, sequence the ITS1 gene, and compare the resulting sequence with globally NCBI database sequences.
MATERIALS AND METHODS
Subjects: this study included 86 people admitted to the dermatology department of (Al-Yarmook teaching hospital) with multiple lesions on different body parts. One of the subjects has lost vision in a single eye. Dermatological examinations of all the subjects showed extensive erythematous plaques with clear borders and scales, scattered red papules with ulceration, and scabs throughout the body. The patients complained of mild itchiness over the lesions and pain in the left eye.
The ethic consent
The consent for the collection of multiple lesions on different parts of the body was obtained from the participants, and also the study was consented to by ethical committees in the Iraqi Ministry of Health/ Al-Yarmook teaching hospital/Baghdad-Iraq)April 2021-August 2021).
Excluded criteria: Patients with diabetes, eye trauma, or other significant medical disorders have been excluded.
DNA extraction
DNA has been extracted from all the samples by using a commercial kit (ZR Fungal/Yeast/Bacterial DNA MiniPrep, Zymo research) .
Amplification of Gene ITS: the amplification of the ITS gene was done by oligonucleotides that specify a region of 600bp a sense primer (ITS F: 5′- TCCGTAGGTGAACCTGCGG -3′) and antisense primer sequence (ITS R:5′ TCCTCCGCTTATTGATATGC-3′) (Xie et al., 2008) Primers supplied by IDT (Canada). The final volume of PCR was 25 µl which included 3µl of eluted DNA, 10 µl PCR Pre Mix (Intron, Korea), 0.5 µl of primer sense and antisense then nuclease-free water was used to complete the volume to 25 µl. After the addition of all the components, the prepared samples were placed into a thermal cycler machine and programmed as follows;: 3 min of denaturation at 94, followed by 35 cycles of 94 °C for 45s, 52°C for 1 min and 72 °C for 1min with final incubation at 72 °C for 7 min using a Thermal Cycler (Gene Amp, PCR system 9700; Applied Biosystems).
RESULTS
The DNA was extracted successfully as the electrophoresis results of the eluted DNA showed a single band by all the samples. After the PCR run, the amplified product also resulted in a single band in electrophoresis at 600 bp for the positive samples, as shown in figure (1; A), and the results showed that 10 out of the 86 samples (11.6%) were positive with Trichophyton interdigitale. The results of electrophoresis are shown in figure (1).
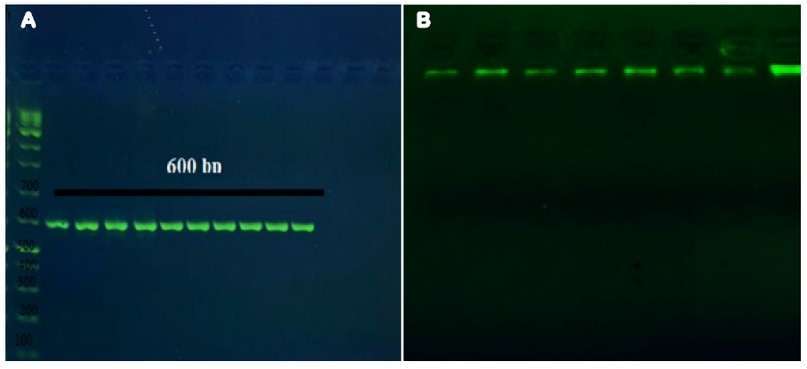
Figure 1. A; PCR product the band size 600 bp. The product was electrophoresis on 2% agarose at 5 volt/cm2. 1x TBE buffer for 1:30 hours. N: DNA ladder (1000 plus).
B; Gel electrophoresis of genomic DNA extraction from fungi, 1.5% agarose gel at 5 vol /cm for 1 hour.
The amplified region of the 10 positive samples has been sequenced by micro gen company (Korea), and each resulted sequence aligned to a high score nucleotides sequence within the BLAST database; the results in figure (2) showed the sequence of the sample (2) aligned to a sequence that shown a high score (568-bit score) and 98% compatibility which accompanied with zero gaps. For further analysis, nucleotides 347bp (corresponding Trichophyton interdigitale isolate AJJM7 internal transcribed spacer gene positions of the GenBank accession no. MH383047) of the ITS gene were used. The sequences were analyzed by using NCBI/BLAST. This study's obtained sequence of ITS gene was deposited in GenBank under accession nos. OL673018.
Summarized details for two types of substitution variations are shown in table (1); sample sequences have been labeled by numbers as shown in the table. The various sequence percentage was 2% for both samples 2 and 5. And this percentage of difference includes 5 variations; two Transition (first one is T\ C located in the 74bp and the second one is A \G located in the 238bp) and three transversions (T\ G in 73bp, T\ G in 72bp and A \C in 235bp). Sample 6 showed variations in two transitions (T\ C in the 50bp and A\G in 207 bp) and three transversions (T\G in 81bp, T\G 83bp and A\C in 210bp). Both samples 7 showed three transversions (at 594 bp G\ T, at 600bp C\ G and 602bp T\ G) while the other samples showed 100% similarity.
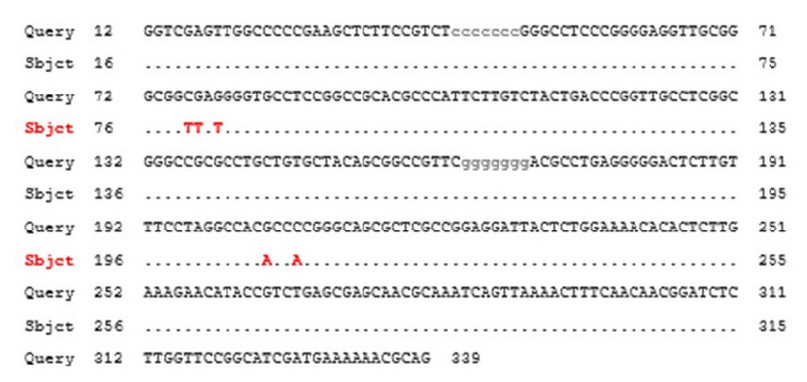
Figure 2. Alignment analysis of ITS1 gene. Query symbolizes the gene sequence of the sample; Subject represents the gene sequence by the database of NCBI.
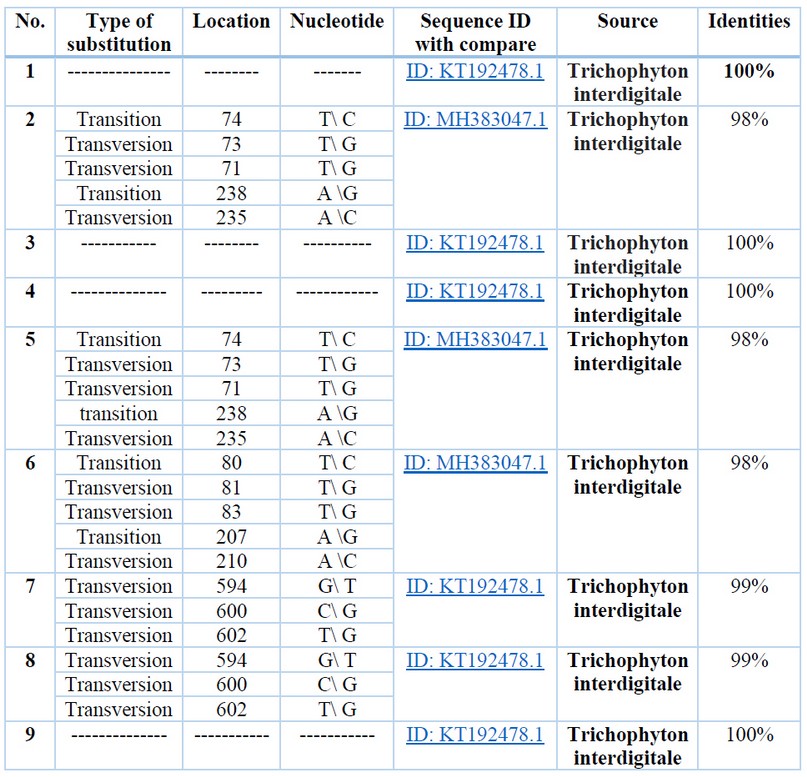
Table 1. Variations of sequences with ITS1 aligned sequence
The phylogenetic tree analysis has been done using the Maximum Likelihood method and is represented in figure (3). The Iraqi isolate of this study (OL673018) is shown in the same clade as the Iranian isolate (KT192478.1), and this clade share 98% similarity with another clade, including France isolate and Germany isolate (MW8980291 and AJ270790.1, respectively) both clades showed 97% similarity with both separate isolates one from Japan and one from the Czech Republic (AB193720.1 and LT897808.1, respectively).
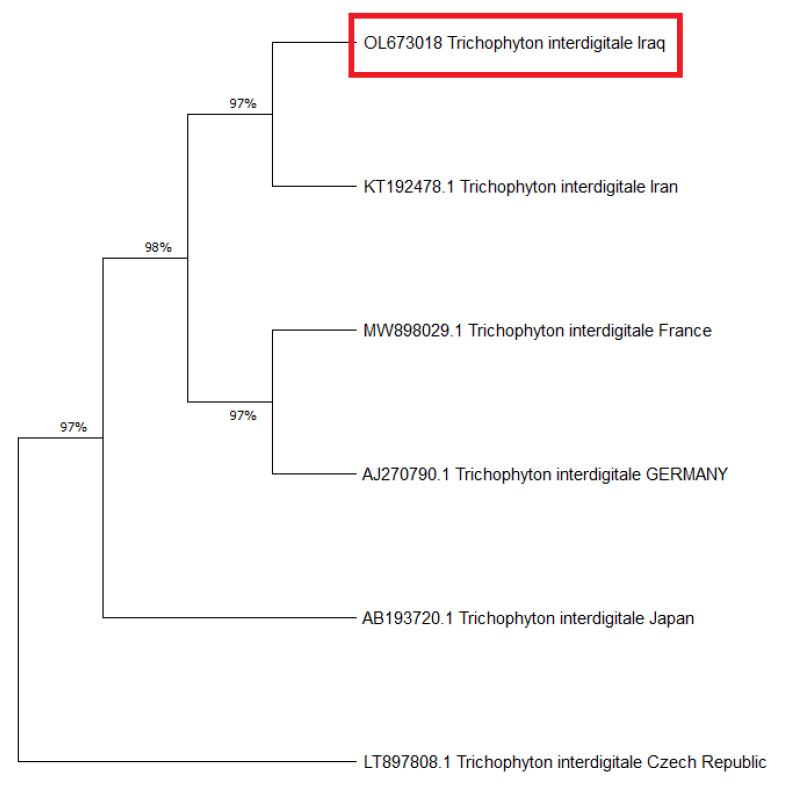
Figure 3. Evolutionary analysis tree by Maximum Likelihood method for the Iraqi isolated in this study with other global isolates
DISCUSSION
Detecting dermatophytes by the usual microscopic methods can cause problems such as false negative results. Due to morphological similarity and existing intermediate forms and variants, unequivocally separating these dermatophytes is not always straightforward, and sampling appropriate isolates for research is often troublesome 9. The molecular study has therefore been used to compare the sequence of the ITS1 gene with the sequences of this gene within the public databases of the globe. For those reasons, this study aimed to diagnose dermatophytes by molecular technique and to compare the resulted sequence of ITS1 gene with the sequences of this gene within the public databases of the globe.
In a previous study by de Hoog et al. 1, many genes were detected and sequenced. And the results revealed a new taxonomy of dermatophytes. T. interdigitale only included the anthropophilic species, and zoophilic T. interdigitale isolates belong to the species T. mentagrophytes. Another study was done by 10 that sequenced four T. mentagrophytes genomes and exhibited that T. mentagrophytes and T. interdigitale belong to the same phylogenetic species. In this study, according to the overall average obtained from intraspecies and interspecies pairwise distances of combination concerning the seven loci, T. interdigitale and T. mentagrophytes were regarded as species complexes that may be affected by the epigenetic change during the localization of the animal and human body. Furthermore, they likely share a common ancestor, and T. interdigitale species are descendants of T. mentagrophytes species. This study confirms the results of the study undertaken by 10. The present study successfully confirmed that ITS1 region is suitable for providing target genes for molecular identification of the Trichophyton interdigitale. Variation in the nucleotide composition of the ITS1 region was successfully employed for recognition among the samples. Various targets have been employed for DNA-based identification and discrimination of pathogenic Trichophyton interdigitale. To the best of our knowledge, this study was one of the first studies that isolated Trichophyton interdigitale and was diagnosed by molecular technique in Iraq. Another study diagnosed Yoshikawa et al., 11 Trichophyton rubum from Al-Dewaniyah General Hospital and found positivity of dermatophyte characters in only 5 clinical samples out of 30. A previous study in Iraq found that most of the organisms isolated from people suspected of dermatophytes belong to the genera Trichophyton 12.
CONCLUSIONS
It was concluded that Trichophyton interdigitale in Iraqi isolates had two types of substitution variations (Transition and Transversion) different than global isolates. Moreover, according to the phylogenetic tree, the similarity was 97% with isolates from the Czech Republic and Japan, whereas 98% with the isolates from France and Germany.
Conflict of interest
The authors declare no conflict of interest.
Acknowledgments
We are thankful to the participants and the Ministry of health /Iraq.
REFERENCES
1. de Hoog GS, Dukik K, Monod M, Packeu A, Stubbe D, Hendrickx M, Kupsch C, Stielow JB, Freeke J, Göker M, Rezaei-Matehkolaei A. Toward a novel multilocus phylogenetic taxonomy for the dermatophytes. Mycopathologia. 2017 Feb;182(1):5-31.
2. Gnat S, Łagowski D, Nowakiewicz A. Genetic predisposition and its heredity in the context of increased prevalence of dermatophytoses. Mycopathologia. 2021 May;186(2):163-76.
3. Wiegand C, Burmester A, Tittelbach J, Darr-Foit S, Goetze S, Elsner P, Hipler UC. Dermatophytosis caused by rare anthropophilic and zoophilic agents. Der Hautarzt; Zeitschrift fur Dermatologie, Venerologie, und verwandte Gebiete. 2019 Aug 1;70(8):561-74.
4. Minnat TR. Epidemiological, Clinical and Laboratory study of Canine Dermatophytosis in Baghdad Governorate, Iraq: Tareq Rifaaht Minnat1 and Jinan Mahmood Khalaf 2. The Iraqi Journal of Veterinary Medicine. 2019 Aug 4;43(1):183-96.
5. Sharma B, Nonzom S. Superficial mycoses, a matter of concern: Global and Indian scenario‐an updated analysis. Mycoses. 2021 Aug;64(8):890-908.
6. Alshehri BA, Alamri AM, Rabaan AA, Al-Tawfiq JA. Epidemiology of Dermatophytes Isolated from Clinical Samples in a Hospital in Eastern Saudi Arabia: A 20-Year Survey. Journal of Epidemiology and Global Health. 2021 Dec;11(4):405-12.
7. Gräser Y, Monod M, Bouchara JP, Dukik K, Nenoff P, Kargl A, Kupsch C, Zhan P, Packeu A, Chaturvedi V, De Hoog S. New insights in dermatophyte research. Medical mycology. 2018 Apr 1;56(suppl_1):S2-9.
8. Ahmadi B, Mirhendi H, Makimura K, de Hoog GS, Shidfar MR, Nouripour-Sisakht S, Jalalizand N. Phylogenetic analysis of dermatophyte species using DNA sequence polymorphism in calmodulin gene. Sabouraudia. 2016 Feb 11;54(5):500-14.
9. Nasrin S, Sina M. Genotyping and Molecular Characterization of Dermatophytes Isolates Collected from Clinical Samples. Archives of Pulmonology and Respiratory Care. 2017 Jun 14;3(1):052-7.
10. Yun-lu GA, Zhi-qing GA, Qiang JU, Min L. Adult tinea capitis and tinea corporis due to Trichophyton violaceum: a case report. Chinese Journal of Mycology. 2015 Oct 28;10(5):297.
11. Yoshikawa FS, Yabe R, Iwakura Y, De Almeida SR, Saijo S. Dectin-1 and Dectin-2 promote control of the fungal pathogen Trichophyton rubrum independently of IL-17 and adaptive immunity in experimental deep dermatophytosis. Innate immunity. 2016 Jul;22(5):316-24.
12. Mohammed SJ, Noaimi AA, Sharquie KE, Karhoot JM, Jebur MS, Abood JR, Al-Hamadani A. A survey of dermatophytes isolated from Iraqi patients in Baghdad City. Al-Qadisiyah Medical Journal. 2015;11(19):10-5.
Received: 19 March 2022 / Accepted: 7 April 2022 / Published:15 Agoust 2022
Citation: Saied Hamied A. Submission and Phylogenetical of Local Isolated Trichophyton interdigitale of Iraqi Patients in NCBI. Revis Bionatura 2022;7(3) 25. http://dx.doi.org/10.21931/RB/2022.07.03.25
