2021.06.01.15
Files > Volume 6 > Vol 6 No 1 2021
INVESTIGATION / RESEARCH
Analysis of Pseudocercospora fijiensis genes upregulated during early interaction with Musa acuminata (var. Dwarf Cavendish)
Bartolomé Chí Manzanero1, Karla Gisel Carreón Anguiano1, Jewel Nicole Anna Todd1, Rufino Gómez Tah1, Rosa Grijalva Arango2, Miguel A. Tzec Simá3, Blondy Canto Canché 1,*
Available from: http://dx.doi.org/10.21931/RB/2021.06.01.15
ABSTRACT
Pseudocercospora fijiensis is a filamentous, hemi[B1] biotrophic fungus whose infection process in banana comprises biotrophic and necrotrophic phases; the biotrophic phase is the longer and less damaging of the two but is nonetheless a crucial stage of fungal establishment in the host. To discover the genes essential in this stage, we conducted an interaction experiment to isolate the transcriptome of the P. fijiensis and Musa acuminata interaction during the first 9 days of infection. Of more than 7000 P. fijiensis genes identified, the fifteen most highly expressed genes (RPKM>500) were analyzed. Specific non-canonical effector candidates were identified following in silico characterization which may be fundamental to pathogenicity. This report reveals essential details of a poorly-elucidated stage of the P. fijiensis-Musa sp. pathosystem.
Keywords: Pseudocercospora fijiensis, transcriptomics, biotrophic stage, upregulated genes, effectors
INTRODUCTION
Banana is the seventh most crucial crop globally, and its production is seriously affected by multiple pests and diseases. One of the most devastating diseases is the Black Leaf Streak disease, commonly known as Black Sigatoka, caused by Pseudocercospora fijiensis. This fungus can cause severe damage to plant foliar tissue, diminishes the plant's photosynthetic capacity, and reduces fruit yields. Fungal infection can result in total plant death, and producers incur substantial losses of up to 100% in some cases1,2.
P. fijiensis is considered the most virulent species of the Sigatoka complex, which comprises the fungi P. musae, P. eumusae and P. fijiensis3. The infection begins when newly released fungal spores contact the banana plant's leaves and germinate on the leaf surface within 2 to 3 hours. Environmental conditions that are wet and humid are required for germination. The resulting germ tube grows epiphytically on the leaf surface for 2 to 3 days before it penetrates the leaf stomata through its formed appendages, the appressoria or stomatopodia4. Other reports have given a faster timeline; in greenhouse conditions, the germ tube formed within 6 hours and penetrated the stomata after 12 to 15 hours5. P. fijiensis has to complete two stages of its infection as a hemibiotrophic fungus: biotrophic and necrotrophic stages. In the biotrophic phase, the pathogen exclusively colonizes the intercellular spaces between mesophyll cells and obtains nutrients from the host apoplast without forming feeding structures called haustoria. The fungus remains in this phase for 3 to 4 weeks before entering the aggressive necrotrophic phase, marked by the appearance of the characteristic black, necrotic spots, then streaks formed on the leaves of the infected plant6.
In recent years, different studies have been done to understand the banana-P better P.fijiensis interaction. Although many omic technologies have been developed, very little is known of what occurs during the plant's early-stage P. fijiensis infection. Gene expression analysis is useful for the identification of genes involved in the plant-pathogen interaction. In one of the earliest studies of P. fijiensis gene expression, cDNA libraries of P. fijiensis grown in different in vitro conditions were compared to identify pathogenicity-related genes in the P. fijiensis-Musa sp. Interaction7. More recently, a group of small, cysteine-rich proteins called effectors has received particular attention in P. fijiensis; several effectors have been predicted: 172 sequences by Arango-Isaza et al., 105 sequences by Chang et al., and 136 sequences by Carreón-Anguiano et al.8,3,9. As a hemibiotroph, it is expected that P. fijiensis would produce effector proteins and toxic secondary metabolites to manipulate the defense response of the host during late interaction, as well as prevent host cell death during the fungal biotrophic phase. Noar and Daub10 studied gene expression data of P. fijiensis in 6-week infected leaves, in comparison with P. fijiensis in vitro monoculture, resulting in interesting and valuable information about the necrotrophic phase of the infection. However, the fungus' repertoire during the biotrophic infection stage is yet to be elucidated. Studying the biotrophic phase of infection is challenging due to the limited quantity of biomass produced by this relatively slow-growing fungus.
In this study, a transcriptome corresponding to the first 9 days of infection of banana cv. Dwarf Cavendish with P. fijiensis was analyzed to uncover pathogen genes that were upregulated at the initial (biotrophic) stage of this plant-pathogen interaction.
MATERIALS AND METHODS
Plant growth conditions
Seedlings were obtained from in vitro culture in the proliferation phase, according to Strosse et al.11. Shoots of 3-4 cm in height, with 2-3 formed leaves were separated and transferred to Murashige and Skoog (MS) medium without growth regulators. The growth conditions were: 1000 to 5000 lux, 25 ± 1oC, and a 16 hours light-8 hours dark photoperiod. After 30 days, plants 7 cm in height were subjected to acclimatization in greenhouse conditions.
Seedlings were removed from containers, roots were cut, and seedlings were planted in trays containing a moist mixture of soil: peat (1: 2 v/v). Plants were maintained for 8 days at 80% relative humidity and watered every 8 days with Hoagland solution12. After 30 days, the plants that reached 15 cm in height and had 5 new leaves were sown in plastic bags with a mixture of soil and peat (2: 1 v/v). After three months, the plants were ready for infection.
Conidiation
Conidia were produced according to Acosta-Suárez et al.13. P. fijiensis strain C1233 was cultivated in 10 Petri dishes with potato dextrose agar (PDA) medium for 14 days and then transferred to 10 flasks, each with 50 ml of liquid potato dextrose broth (PDB) for an additional 14 days to obtain sufficient biomass. Mycelium was harvested, finely macerated in aseptic conditions and inoculated on PDA agar slants. Cultures were incubated in constant white light at 20⁰C for 18 days until a fine grayish layer was observed. The presence of conidia was confirmed by microscopic observation.
Infection and sampling
Conidia were harvested and quantified using a Neubauer chamber. The concentration was adjusted to 105 conidia ml-1 with 1% gelatin for plant infection. The banana plants were inoculated on the abaxial surface of the 2nd and 3rd leaves with the conidial solution. For control plants, the leaves were inoculated with a 1% gelatin solution. Sample collection was carried out on days 0, 3, 6, and 9 post-inoculation (3 plants per data point). Samples were packaged, labeled, and frozen in liquid nitrogen until extraction.
Total RNA extraction and transcriptome sequencing
RNA was extracted independently from each sample (0.5 g) using Concert™ Plant RNA reagent (Invitrogen), following the manufacturer's instructions. For transcriptomics analysis, a pool of all data points (5 µg of RNA from each point) was prepared, i.e., each pool contained the biological replicates corresponding to samples 0, 3, 6, and 9 d post-infection. In total, there were 6 pools: 3 for control samples and 3 for treatment.
Samples were sequenced at IBT facility (UNAM, Mexico) on the Illumina NextSeq platform with paired-end sequencing for 2×75 cycles.
Data analysis
The raw transcriptome data quality assessment was done using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and fast trimmer to retain reads at least 75 bp long with a minimum Phred quality score of 30. Clean reads were obtained by removing the empty reads, adaptor sequences, and the low-quality sequences (reads with unknown base pairs "N"). Once verified, the reads were mapped against the reference genome of P. fijiensis strain CIRAD86, and the annotation data for P. fijiensis genes was downloaded from JGI genome portal of P. fijiensis (https://mycocosm.jgi.doe.gov/Mycfi2/Mycfi2.home.html). To map the reads obtained from Musa acuminata, we used the genome reported in Banana Genome Hub (https://banana-genome-hub.southgreen.fr/). Alignment of RNA Seq data (in BAM format) was done using SMALT. An in-house python script was used to make in tandem modifications to the data. Each sequencing library had at least 10 million reads per sample.
To identify the P. fijiensis genes with the highest expression during biotrophy, we normalized the quantification of the data, i.e., the mean gene expression of each sample was expressed as reads per kilobase per million reads sequenced (RPKM). Using the formula "RPKM = (read count * 10^9) / (read length * total read count), the genes with the highest value of RPKM were selected. Protein identifications and functional predictions were obtained by searching for sequence homologs using BlastP (E-Value 1.0E-10) at GenBank; if an uncharacterized protein represented the most significant match in BlastP, the first match in the list of homologous proteins where a protein function was available was considered. The taxonomic distribution of homologs was established from BlastP results. Homology with pathogenesis-related genes was determined by BLASTing in the Pathogen-Host Interaction (PHI) database (http://phi-blast.phi-base.org/).
The computation of theoretical molecular weight (Mw), amino acid composition, and cysteine content were performed using the ExPASy Compute tool (https://web.expasy.org/). The presence of a signal peptide was predicted using SignalP 4.1 (http://www.cbs.dtu.dk/services/SignalP/index.php). Information about the subcellular protein localization was obtained using WolfPsort (https://wolfpsort.hgc.jp/) and LOCALIZER14. Effector prediction was conducted with EffectorP 2.0 (http://effectorp.csiro.au/). Domains were searched in the InterProscan database (https://www.ebi.ac.uk/interpro/search/sequence/), and motifs using MEME suite (http://meme-suite.org/tools/meme).
RESULTS AND DISCUSSION
Three libraries, each containing more than 10 million reads, were obtained. In the infected samples, 82-86% of sequences mapped to M. acuminata while 3.8-7.8 % mapped to P. fijiensis (Table 1).
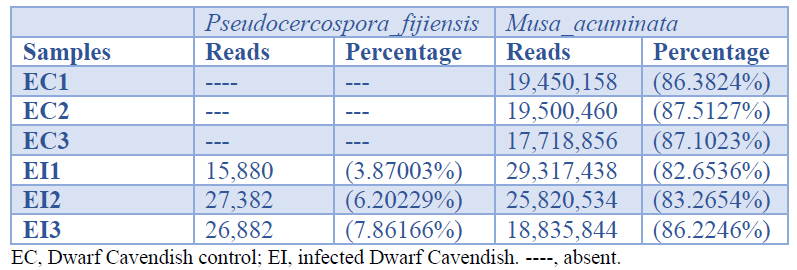
Table 1. Number and percentage of sequences mapped with the genome of Musa acuminata and Pseudocercospora fijiensis
In total, 7144 genes of P. fijiensis were identified. Paying attention to those genes with the highest expression, we found 40 genes with RPKM>200, and among these, nine with RPKM >1000 and two with RPKM >3000 (Figure 1). Table 2 lists, in descending order, the 15 genes with the highest expression identified during P. fijiensis biotrophy, while Table 3 corresponds to the in silico characterization of these genes.
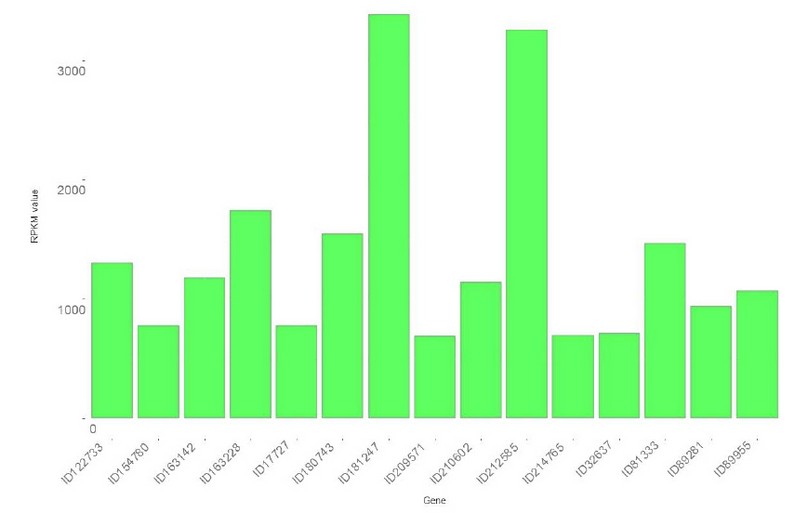
Figure 1. Top 15 genes upregulated during the biotrophic stage of Pseudocercospora fijiensis infection of Musa acuminata cv Dwarf Cavendish.
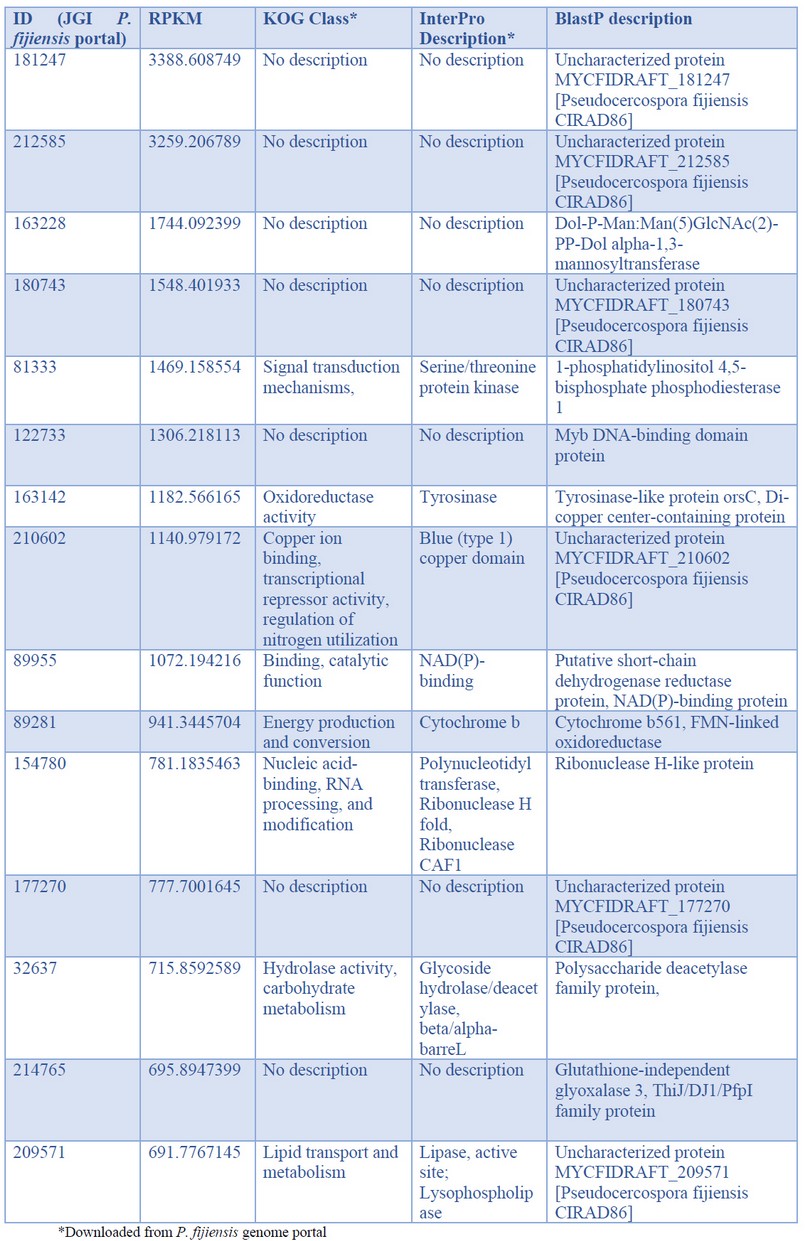
Table 2. Pseudocercospora fijiensis genes with the highest expression values during biotrophy phase infection of banana cv. Dwarf Cavendish
The first two genes with the highest expression were codified for small proteins with unknown function (Tables 2 and 3). Searching in the PHI database, both genes had a single hit respectively: P. fijiensis protein 181247 matched with the trpA gene of Aspergillus fumigatus and the protein 212585 with PKS1 of Alternaria alternata. However, both queries match the subjects on short fragments; protein 181247 shares 45% identity in 30 of the 517 amino acids of the trpA protein, while protein 212585 shares 27% identity in 55 of the 1484 amino acids of the PKS1 protein. These genes are also expressed during the necrotrophic phase of P. fijiensis infection, but with much lower expression than found here during biotrophy, especially protein 212585 (Table S1 in Noar and Daub10). Therefore, although their functions are mostly unknown, it is possible to hypothesize that these proteins play essential roles in the pathogenesis process of P. fijiensis, principally during the biotrophic stage. The short fragments of the queries matching the subjects were not enough to retrieve these genes when BLASTed against the GenBank database. BlastP only retrieved one subject in each case, uncharacterized proteins of P. fijiensis CIRAD86. These upregulated genes are, therefore, specific to this pathogen.
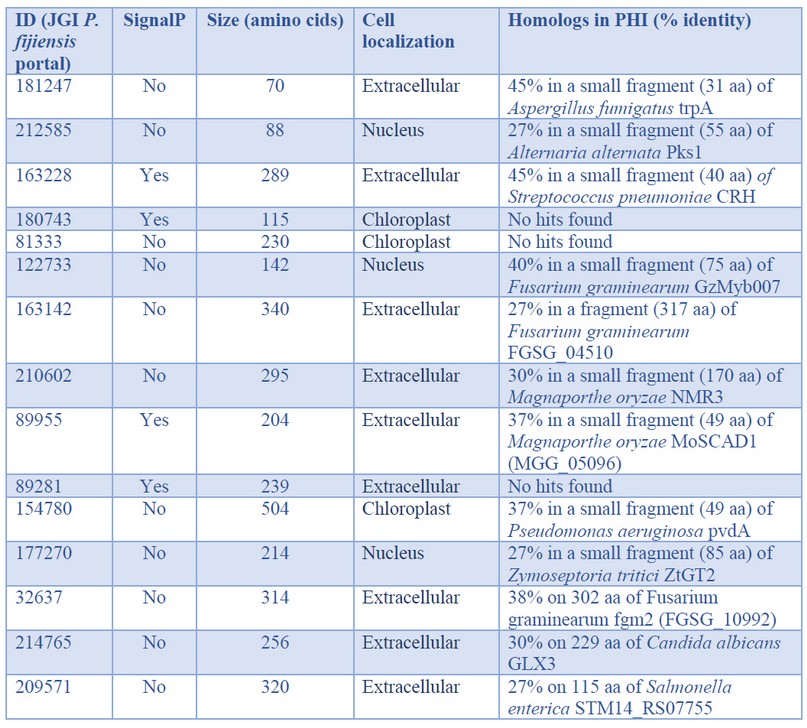
Table 3. In silico characterization of the P. fijiensis genes with the highest expression observed during biotrophy phase infection of banana cv. Dwarf Cavendish
The third-highest expressed gene of P. fijiensis in the transcriptome corresponded to a Dol-P-mannosyltransferase. Recently, Pham et al.15 studied the biosynthetic machinery of the cell wall of the obligate biotrophic Blumeria graminis f. sp. hordei, and found that one of the most highly expressed enzymes was a Dol-P-mannosyltransferase. This economically important pathogen of barley has large amounts of mannosyl residues in its cell wall compared to other fungi; so, the high expression of enzymes involved in the synthesis of N-glycans in B. graminis f. sp. hordei is congruent. The high expression of the Dol-P-mannosyltransferase in P. fijiensis suggests that mannosyl residues play an essential role in the biotrophic stage of this fungus. During this stage, P. fijiensis needs to remain undetected by the host to complete the infection process. Part of the fungus' survival strategy is to maintain its cell wall, preventing the detection of cell wall fragments by host receptors that trigger plant defense responses that are detrimental to the pathogen.
Protein families related to oxidoreduction are involved in pathogenesis, secondary metabolite biosynthesis, and toxins or drugs' detoxification. Tyrosinases are enzymes with oxidoreductase activity and are also virulence factors of necrotrophic fungi16,17. One of the most highly expressed proteins found in our analysis was a tyrosinase. Similarly, a tyrosinase was an enriched term in the transcriptome of the biotrophic fungus Moniliophthora perniciosa during Theobroma cacao18, and during the biotrophic phase of tomato and potato infection by Phytophthora infestans19. The other highly expressed redox protein found in the P. fijiensis transcriptome was a NADPH-binding short-chain dehydrogenase (ID 89955). It appears that short-chain dehydrogenases are under different temporal regulation in P. fijiensis since during P. fijiensis necrotrophy, 11 upregulated short-chain dehydrogenases were found, most likely involved in secondary metabolism10.
Similarly, a short-chain dehydrogenase was upregulated specifically during the biotrophic phase of Colletotrichum graminícola20, which supports this enzyme's finding in P. fijiensis biotrophy. However, the role of tyrosinases and short-chain dehydrogenases during the fungal biotrophy stage remains to be determined. Other identified proteins involved in redox activity were a blue copper domain-containing protein (ID 210602) and a cytochrome b561 domain-containing protein (ID 89281), demonstrating the importance of electron transport and redox homeostasis in the P. fijiensis biotrophic stage.
One of the upregulated genes codifies a glyoxalase 3 enzyme (GLY3), which catalyzes methylglyoxal (MG) conversion to D-lactate in a glutathione-independent reaction. MG is produced during various abiotic stress conditions such as salinity, drought, flooding, cold, heat, etc., as well as during biotic stresses. The plants possess multiple DJ-1/PfpI GLY3 to detoxify MG, e.g., Zea mays have eleven GLY3, Sorghum bicolor has five and Musa acuminata has four21. In fungi, the first reports of identification of GLY3 were in Saccharomyces cerevisiae and Candida albicans, where GLY3 was associated with stress tolerance22. To the best of our knowledge, the present work is the first report of DJ-1/PfpI GLY3 in fungal biotrophy, and it suggests that the detoxification of MG is one important adaptive strategy of P. fijiensis to colonize banana.
For all pathogens, efficient nutrition is a prerequisite for successful establishment. Among the upregulated genes during biotrophy, there were genes involved in the uptake of purines from the host environment (ID 64453), lipid transport (ID 209571), and carbohydrate hydrolysis (ID 32637). Usually, purine uptake is low in the fungal biotrophic stage, fueled by de novo biosynthesis23,24. In another P. fijiensis study, adenylosuccinate synthetase (AdSS) and orotidine 5′-phosphate decarboxylase (OMP decarboxylase), key enzymes involved in de novo biosynthesis of purines and pyrimidines, respectively, were identified in the cell wall proteomes of the highly virulent Oz2b strain, signaling that these de novo biosynthetic pathways are important for virulence of this pathogen25. Here, another highly expressed enzyme during biotrophy was a putative purine permease, revealing that both routes are functional in P. fijiensis. Other fungi are also versatile and can take up nucleosides from the host and utilize nucleosides through their salvage metabolism or de novo pathways26.
Interestingly, one of the top-expressed genes in P. fijiensis is a predicted ribonuclease H-like protein. The phylogenetically-related fungus, Zymoseptoria tritici, secretes the ribonuclease effector Zt6 during wheat infection. The characterization of Zt6 revealed toxicity against bacteria, yeasts, filamentous fungi, and phytotoxicity on tobacco and wheat27. Ribonuclease H-like proteins comprise a superfamily in the obligate biotroph, Blumeria graminis, in which they are termed RALPHs; it is the most extensive set of secreted effectors in this pathogen. Pennington et al.28 demonstrated that the RALPH effector protein CSEP0064/BEC1054 interferes with the degradation of host ribosomal RNA and represses plant immunity. Since the ribonuclease H-like protein (ID 154780) is upregulated during the biotrophic stage in P. fijiensis, it is tempting to speculate that it plays a similar role in the black Sigatoka disease as the B. graminis CSEP0064/BEC1054 effector in powdery mildew disease on cereals and grasses. However, its role in antimicrobial competition and niche protection cannot be ruled out.
When BLASTing in the PHI database, most of the genes retrieved hits, but poor coverage and identity with their respective hits were observed except genes 163142 and 32637. Three genes had no hits at all (Table 3). Low conservation in pathogenicity-related proteins, especially effector proteins, is quite common.
Effectors play fundamental roles in pathogenesis, playing diverse roles in camouflaging the pathogen, protecting the pathogen from host defense mechanisms, suppressing said host defenses, or attacking host cells. Effector identification can be difficult; in most cases, effectors do not share homology with other known proteins. Furthermore, not all effectors present with the same characteristics, although a few have been established to identify identification better. Canonical effectors are predicted based on the small size (<400 amino acids), richness in cysteine (>4 Cys or >2% per sequence), the occurrence of a signal peptide, and absence of a transmembrane domain9. Other criteria to identify in silico effectors include: high in planta expression, pathogenesis-related domains, and discontinuous taxonomic distribution29. Besides, many effectors have unknown functions, with no homologs in other fungi, or are homologous only to closely-related organisms. To explore whether hypothetical proteins of P. fijiensis with unknown functions are probable effectors, they were further analyzed in silico. Table 4 presents the possible effector candidates found in the P. fijiensis early infection transcriptome. The two proteins found to be related to fungal effectors were included in this list.
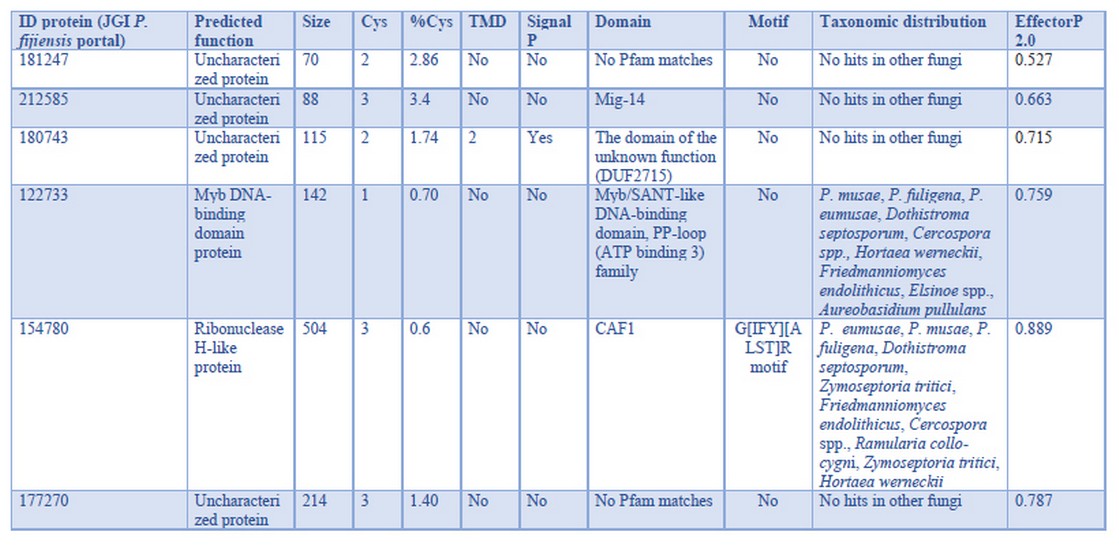
Table 4. P. fijiensis effector candidates with full expression during biotrophy on banana cv. Dwarf Cavendish
Restricted phylogenetic distribution in these genes (Table 4) is consistent with what is expected for effectors30. The ribonuclease H-like protein and the Myb DNA-binding domain TF have discontinuous taxonomic distribution; meanwhile, the other four genes are exclusive to P. fijiensis, i.e., no hits in other fungi. None of the six effector candidates meet canonical characteristics; thus, these genes are probably non-canonical effectors. The protein 212585 matches the Mig-14 Pfam domain, which is present in bacterial proteins and contributes to bacterial resistance to antimicrobial peptides31. This effector could enable P. fijiensis to cope with host antimicrobials or to compete inside the host against microbial endophytes to conquer the niche.
Like the proteins mentioned above, the proteins with IDs 181247 and 177270 are unique to P. fijiensis, but have no identifiable domains. Many effectors have shown to be unique to their pathogens as they evolve in specific interactions with the host.
In the motif analysis, only the protein corresponding to ID 154780 contained a motif: the G[IFY][ALST]R motif (p-value of 4.35e-05), which has been found in secreted pathogenicity-related proteins expressed in the haustoria of the biotroph Flax rust and effector proteins secreted by Puccinia triticina32,33,34, another biotrophic fungus. Therefore, this motif in protein 154780 is consistent with its role in the biotrophic stage.
Modulation of gene expression requires the participation of transcription factors (TFs). The protein 122733 contains a Myb DNA-binding domain, which is a large and ubiquitous family of TFs. In Fusarium graminearum, the mutation of the nuclear protein MYT3, a Myb-Like TF reduced pathogenicity35. Another putative TF was identified among the top 40 genes with the highest expression. This protein (ID 213137) is a nucleic acid-binding protein, a putative zinc finger C2H2-type TF. Zinc-finger proteins (ZNFs) are ubiquitous proteins able to interact with DNA, RNA, and other proteins. ZNFs are involved in some processes such as transcriptional regulation, ubiquitin-mediated protein degradation, signal transduction, DNA repair, and cell migration. In the fungus, Ustilago maydis, the transcription factor Mzr1, a Cys2His2-type zinc finger, regulates fungal gene expression during the biotrophic growth stage36. Bioinformatic characterization of the protein 213137 predicts nuclear localization, which supports that protein 213137 plays a similar role in P. fijiensis as does Mzr1 in U. maydis.
CONCLUSION
This is the first report that focuses on the identification and in silico characterization of genes involved in the biotrophic stage of infection of P. fijiensis, the causative agent of the black Sigatoka disease of banana. Our transcriptomic analysis successfully uncovered P. fijiensis proteins involved in repressing host defense immunity and coping with environmental conditions inside the host. Six of the upregulated genes are effector candidates, three of them with novel, unknown functions.
Acknowledgments
This work was supported by the National Council of Science and Technology (CONACYT) project FOSEC-SEP 220957, Mexico. Fellowship from CONACyT- 589301 for K. G. Carreón-Anguiano, CONACyT-291236 for J. N. A. Todd, CONACyT-702045 for R. Gómez Tah and the support by BioAli-CYTED are acknowledged. Authors thank CIBBE-ESPOL (Ecuador) by financial support for this Open Access publishing.
REFERENCES
- Alakonya AE, Kimunyeb J, Mahukuc G, Amaha D, Uwimanab B, Brownd A, et al. Progress in understanding Pseudocercospora banana pathogens and the development of resistant Musa germplasm. Plant Pathol. 2018;67:759–770.
- Churchill ACL. Mycosphaerella fijiensis, the black leaf streak pathogen of banana: progress towards understanding pathogen biology and detection, disease development, and the challenges of control: M. fijiensis, BLS pathogen of banana. Mol Plant Pathol. 2011;12(4):307–28.
- Chang TC, Salvucci A, Crous PW, Stergiopoulos I. Comparative Genomics of the Sigatoka Disease Complex on Banana Suggests a Link between Parallel Evolutionary Changes in Pseudocercospora fijiensis and Pseudocercospora eumusae and Increased Virulence on the Banana Host. PLoS Genet. 2016; 12(8): e1005904.
- Álvarez JC, Rodriguez HA, Rodriguez-Arango E, Monsalve ZI, Morales O. JG, Arango I RE. Characterization of a differentially expressed phenylalanine ammonia-lyase gene from banana induced during Mycosphaerella fijiensis infection. J Plant Stud. 2013;2(2):35.
- Rodríguez HA, Rodriguez-Arango E, Morales JG, Kema G, Arango RE. Defense gene expression associated with biotrophic phase of Mycosphaerella fijiensis M. morelet infection in banana. Plant Dis. 2016;100(6):1170–5.
- Lepoivre P, Busogoro JP, Etame JJ, El Hadrami A, Carlier J, Harelimana G, et al. Mycosphaerella leaf spot diseases of bananas: present status and outlook [Internet]. Bioversityinternational.org. [cited 2020 15 June]. Available from: https://www.bioversityinternational.org/e-library/publications/detail/mycosphaerella-leaf-spot-diseases-of-bananas-present-status-and-outlook/
- Cho Y, Hou S, Zhong S. Analysis of Expressed Sequence Tags from the Fungal Banana Pathogen Mycosphaerella fijiensis. Open Mycol J. 2008;2:61-73.
- Arango-Isaza RE, Diaz-Trujillo C, Dhillon B, Aerts A, Carlier J, Crane CF, et al. Combating a global threat to a clonal crop: Banana black Sigatoka pathogen Pseudocercospora fijiensis (synonym Mycosphaerella fijiensis) genomes reveal clues for disease control. PLoS Genet. 2016;12(8):e1005876.
- Carreón-Anguiano KG, Islas-Flores I, Vega-Arreguín J, Sáenz-Carbonell L, Canto-Canché B. EffHunter: A tool for prediction of effector protein candidates in fungal proteomic databases. Biomolecules. 2020;10(5):712.
- Noar RD, Daub ME. Transcriptome sequencing of Mycosphaerella fijiensis during association with Musa acuminata reveals candidate pathogenicity genes. BMC Genomics. 2016;17(1):690.
- Strosse H, Van den Houwe I, Panis B. Banana Cell and Tissue Culture - Review. Banana improvement: cellular, molecular biology, and induced mutations. Catholic University of Leuven. 2004.1-12.
- Hoagland DR. The water-culture method for growing plants without soil [Internet]. Rev. ed. / by D.I. Arnon. Berkeley, Calif: College of Agriculture, University of California; 1950. Available from: https://openlibrary.org/books/OL25240089M.opds.
- Acosta-Suárez M, Alvarado-Capó Y, Cruz-Martín M, Leyva-Mora M, Roque-Morales B. Evaluación en casa de cultivo de la respuesta a la Sigatoka negra de dos cultivares de Musa mediante la inoculación artificial de suspensiones conidiales de Pseudocercospora fijiensis. Biotecnol Veg. 2004;4(2). Available from: https://revista.ibp.co.cu/index.php/BV/article/view/386/html.
- Sperschneider J, Catanzariti A-M, DeBoer K, Petre B, Gardiner DM, Singh KB, et al. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep. 2017;7(1):44598.
- Pham TAT, Kyriacou BA, Schwerdt JG, Shirley NJ, Xing X, Bulone V, et al. Composition and biosynthetic machinery of the Blumeria graminis f. sp. hordei conidia cell wall. Cell Surf. 2019;5(100029):100029.
- Heard S, Brown NA, Hammond-Kosack K. An Interspecies Comparative Analysis of the Predicted Secretomes of the Necrotrophic Plant Pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS One. 2015;10(6):e0130534.
- Wang X, Jiang N, Liu J, Liu W, Wang G-L. The role of effectors and host immunity in plant-necrotrophic fungal interactions. Virulence. 2014;5(7):722–32.
- Teixeira PJPL, Thomazella DP de T, Reis O, do Prado PFV, do Rio MCS, Fiorin GL, et al. High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell. 2014;26(11):4245–69.
- Kagda MS, Martínez-Soto D, Ah-Fong AMV, Judelson HS. Invertases in Phytophthora infestans localize to haustoria and are programmed for infection-specific expression. MBio. 2020;11(5):e01251-20.
- Torres MF, Ghaffari N, Buiate EAS, Moore N, Schwartz S, Johnson CD, et al. A Colletotrichum graminicola mutant deficient in the establishment of biotrophy reveals early transcriptional events in the maize anthracnose disease interaction. BMC Genomics. 2016;17(1):202.
- Ghosh A, Kushwaha HR, Hasan MR, Pareek A, Sopory SK, Singla-Pareek SL. Presence of unique glyoxalase III proteins in plants indicates the existence of shorter route for methylglyoxal detoxification. Sci Rep. 2016;6(1):18358.
- Wilson MA. Metabolic role for yeast DJ-1 superfamily proteins. Proc Natl Acad Sci U S A. 2014;111(19):6858–9.
- Chitty JL, Fraser JA. Purine acquisition and synthesis by human fungal pathogens. Microorganisms. 2017;5(2):33.
- Fernandez J, Yang KT, Cornwell KM, Wright JD, Wilson RA. Growth in rice cells requires de novo purine biosynthesis by the blast fungus Magnaporthe oryzae. Sci Rep. 2013;3(1):2398.
- Burgos-Canul YY, Canto-Canché B, Berezovski MV, Mironov G, Loyola-Vargas VM, Barba de Rosa AP, et al. The cell wall proteome from two strains of Pseudocercospora fijiensis with differences in virulence. World J Microbiol Biotechnol. 2019;35(7):105.
- Daumann M, Golfier P, Knüppel N, Hahn M, Möhlmann T. Botrytis cinerea can import and utilize nucleosides in salvage and catabolism and BcENT functions as high affinity nucleoside transporter. Fungal Biol. 2016;120(8):904–16.
- Kettles GJ, Bayon C, Sparks CA, Canning G, Kanyuka K, Rudd JJ. Characterization of an antimicrobial and phytotoxic ribonuclease secreted by the fungal wheat pathogen Zymoseptoria tritici. New Phytol. 2018;217(1):320–31.
- Pennington HG, Jones R, Kwon S, Bonciani G, Thieron H, Chandler T, et al. The fungal ribonuclease-like effector protein CSEP0064/BEC1054 represses plant immunity and interferes with degradation of host ribosomal RNA. PLoS Pathog. 2019;15(3):e1007620.
- Liang P, Liu S, Xu F, Jiang S, Yan J, He Q, et al. Powdery mildews are characterized by contracted carbohydrate metabolism and diverse effectors to adapt to obligate biotrophic lifestyle. Front Microbiol. 2018;9:3160.
- Giraldo MC, Valent B. Filamentous plant pathogen effectors in action. Nat Rev Microbiol. 2013;11(11):800–14.
- Brodsky IE, Ernst RK, Miller SI, Falkow S. mig-14 is a Salmonella gene that plays a role in bacterial resistance to antimicrobial peptides. J Bacteriol. 2002;184(12):3203–13.
- Catanzariti A-M, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18(1):243–56.
- Kamel L, Tang N, Malbreil M, San Clemente H, Le Marquer M, Roux C, et al. The comparison of expressed candidate secreted proteins from two arbuscular mycorrhizal fungi unravels common and specific molecular tools to invade different host plants. Front Plant Sci. 2017;8:124.
- Zhang Y, Wei J, Qi Y, Li J, Amin R, Yang W, et al. Predicating the effector proteins secreted by Puccinia triticina through transcriptomic analysis and multiple prediction approaches. Front Microbiol. 2020;11.
- Kim Y, Kim H, Son H, Choi GJ, Kim J-C, Lee Y-W. MYT3, a Myb-like transcription factor, affects fungal development and pathogenicity of Fusarium graminearum. PLoS One. 2014;9(4):e94359.
- Zheng Y, Kief J, Auffarth K, Farfsing JW, Mahlert M, Nieto F, et al. The Ustilago maydis Cys2His2-type zinc finger transcription factor Mzr1 regulates fungal gene expression during the biotrophic growth stage. Mol Microbiol. 2008;68(6):1450–70.
Received: 23 November 2020
Accepted: 14 January 2021
Bartolomé Chí Manzanero1, Karla Gisel Carreón Anguiano1, Jewel Nicole Anna Todd1, Rufino Gómez Tah1, Rosa Grijalva Arango2, Miguel A. Tzec Simá3, Blondy Canto Canché 1,*
1Unidad de Biotecnología, Centro de Investigación Científica de Yucatán, A.C., Calle 43 No. 130 X 32 y 34, Col. Chuburná de Hidalgo, C.P. 97205 Mérida, Yucatán, México.
2Unidad de Recursos Naturales, Centro de Investigación Científica de Yucatán, A.C., Calle 43 No. 130 X 32 y 34, Col. Chuburná de Hidalgo, C.P. 97205 Mérida, Yucatán, México.
3Unidad de Boquímica y Biología Molecular de Plantas, Centro de Investigación Científica de Yucatán, A.C., Calle 43 No. 130 X 32 y 34, Col. Chuburná de Hidalgo, C.P. 97205 Mérida, Yucatán, México.
*Correspondence: [email protected]; Tel.: +52-999-942-8330
