CS 2018.01.01.5
Files > Conference Series > 2018 > CIBB 2018
Bionatura Conference Series Vol 1. No 1. 2018
Publicación del IV CONGRESO INTERNACIONAL DE BIOTECNOLOGÍA Y BIODIVERSIDAD IV CIBB y XV FORO INTERNACIONAL BANANERO
INVESTIGATION / RESEARCH
Phosphorylation Analysis of the starch synthase IV-Arabidopsis thaliana SSIV-CR region.
Análisis de fosforilación de la región SSIV-CR de almidón sintasa IV- en Arabidopsis thaliana.
Cristina Calderón1, Ángel Mérida2.
Disponible en: http://dx.doi.org/10.21931/RB/CS/2018.01.01.5
ABSTRACT
Starch synthase (SSIV) plays a specific role in starch synthesis. Previous evidence in our laboratory has shown that SSIV forms dimers, which depends on a region located between the extended coiled-coil region and the glucosyltransferase domain of SSIV. This region is highly conserved between all SSIV enzymes sequenced to date. In this work, we have analyzed the potential phosphorylation of two amino acid residues of the protein (tyrosine 515 and serine 459) and its possible role in the dimerization and activity of SSIV. We have obtained mutated versions of the AtSSIV polypeptide where the amino acid Tyr 515 has been changed to Phe or Glu residue and Ser 459 has been changed to Ala or Asp. We show that the phosphorylation of the Tyr 515 would prevent the formation of a dimer and hence the interaction with FBN1b and suggest that the change of the Tyr 515 residue by a Phe does not affect to the formation of a dimer but reduces the enzymatic activity of the protein considerably. A similar decrease was observed when the amino acid was changed to a Glu residue, suggesting that Tyr 515 residue is necessary for the AtSSIV activity.
Keywords: phosphorylation, protein-protein interaction, starch synthase IV, starch initiation.
RESUMEN
La enzima almidón sintasa (SSIV) juega un papel específico en la síntesis de almidón. Evidencias previas de nuestro laboratorio ha demostrado previamente que SSIV forma dímeros, que dependen de una región situada entre la región Long coiled-coil y el dominio glucosiltransferasa de la SSIV. Esta región está altamente conservada entre todas las enzimas SSIV secuenciadas hasta la fecha. En este trabajo, hemos analizado la posible fosforilación de dos residuos de aminoácidos de la proteína (tirosina 515 y serina 459) y su posible papel en la dimerización y la actividad de la SSIV. Hemos obtenido versiones mutadas del polipéptido AtSSIV donde el aminoácido Tyr 515 se ha cambiado por un residuo de Phe o Glu y la Ser 459 se ha cambiado por Ala o a Asp. Mostramos que la fosforilación de Tyr 515 evitaría la formación de un dímero y, por lo tanto, la interacción con FBN1b, y sugiere que el cambio del residuo Tyr 515 por un Phe no afecta a la formación de un dímero, pero reduce considerablemente la actividad enzimática de la proteína. Se observó una disminución similar cuando el aminoácido se cambió a un residuo de Glu, sugiriendo que el residuo de Tyr 515 es necesario para la actividad de AtSSIV.
Palabras clave: fosforilación, interacción proteína-proteína, almidón sintasa IV, iniciación del almidón.
INTRODUCTION
Starch is a polysaccharide with unique physical and chemical properties 1. Starch synthesis and accumulation takes place in plastid in terrestrial plants and green algae, whereas it is accumulated in the cytoplasm of red algae and glaucophytes 1, 2. Starch forms water-insoluble granules formed by two polysaccharides: amylose, and amylopectin3. The amylopectin molecules adopt a radial orientation in the starch granules, with the reducing extreme oriented to the center of the granule and the non-reducing ends situated at the external surface, from which the elongation of the glucose chains is made 4.
The synthesis of starch requires the concerted action of different enzymatic activities: elongation of chains (starch synthases), branching (starch branching enzymes) and debranching (isoamylases, necessary to eliminate some of the branches originated by the branching enzymes) 4. Five different classes of starch synthases are found in all plants, four soluble starch synthase (SS) classes SSI, SSII, SSIII and SSIV, and the granule-bound starch synthase (GBSSI) 5. In several species, for example, Arabidopsis thaliana, there is a single copy of these enzymes 6. The SSI-SSIV elongate amylopectin chains 1. All the SSs display three regions: a chloroplast transit peptide (CTP), a carboxy-terminal domain with high homology to the glycogen synthase (GS), and an amino-terminal domain specific of each enzyme. The catalytic domain of SSs resides in the conserved carboxy-terminal region. In this conserved region can be found a starch catalytic domain (GT-5) (Pfam PF08323) and a glucosyltransferase domain-1 (GT-1) (Pfam PF00534) 7, 8.
Several investigations have demonstrated that the amino-terminal region could be responsible for substrate interaction as well as enzyme complexes formation, interacting with other enzymes involved in starch synthesis 7, 9. Several studies in A. thaliana have shown that SSIV controls the number of starch granules synthesized in the chloroplast. Its elimination determined the reduction of starch content and the accumulation of only one or two granules per chloroplast 10, which are bigger than those found in WT plants. Besides, there are indications that SSIV participates in the formation of the primer required for the de novo synthesis of a starch granule 5, 9. The N-terminal part of SSIV displays four domains with coiled-coils structure (long coiled-coils regions, LCC). Between the LCC regions and the C-terminal domain of SSIV can be observed a stretch of amino acids without a specific structure that is highly conserved among the different SSIV sequenced to date (CR region). Our group has demonstrated that the LCC regions are involved in the interaction of SSIV with fibrillins 1a and 1b (FBN1s). Those fibrillins are two of the main components of plastoglobules, lipoproteins particles associated to the thylakoid membranes 11. The interaction SSIV-FBN1s, mediated by the LCC domains, determines the dot-like pattern of expression of SSIV in the chloroplast. This group has also demonstrated that SSIV forms a homodimer in vivo. The CR domain is necessary for the dimerization of the protein, and the dimerization seems to be necessary for the activity of the protein. At the same time, results obtained by this Group suggest that it is the AtSSIV-AtSSIV dimer the form that interacts with the fibrillins 1. It is interesting to note the presence of a highly conserved tyrosine residue in the CR domain. This residue displays a high probability of being phosphorylated (NetPhos2.0, www.cbs.dtu.dk/services/NetPhos/). Also, close to the CR domain there is a Ser residue that also shows a high probability to be phosphorylated 12. In the present study, we analyze the potential phosphorylation of the tyrosine and serine residues cited above and its possible role in the dimerization and activity of SSIV.
MATERIALS AND METHODS
Plant Materials- Nicotiana benthamiana plants were grown in soil pots in a greenhouse under a 16-h light/ 8-h dark cycle at 22°C.
Escherichia coli strains: DH5α strain was cultured in Petri plates with 30 mL solid Luria-Bertani (LB) medium containing 50 µg/mL of ampicillin (resistance of the pJET1.2/blunt vector), 50 µg/mL of gentamycin (pDONR207 vector), 100 µg/mL of kanamycin (pEarleyGate vector), or 25 µg/mL of spectinomycin (pXCGW-(cCFP), pXNGW-(nYFP)) vectors. Cultures were incubated at 37°C.
E. coli strain BL21 (DE3) ∆glgCAP was inoculated in sterile tubes with 50 mL Luria-Bertani (LB) liquid medium containing 50 µg/mL of kanamycin (endogenous resistance of this strain) and 50 µg/mL of ampicillin (when transformed with pDEST17 or pET45b (+) vectors).
Plasmid construction- The different mutated versions of SSIV (with and without the chloroplast transit peptide) were amplified by the overlapping PCR technique using the specific oligonucleotides. Resulting fragments were cloned into pJET1.2 vector using the T4 DNA ligase and transformed into the DH5α Escherichia coli strain by the heat-shock method 13. Plasmids were isolated using the Isolate II Plasmid Mini kit (BIOLINE) and sent to be sequenced by STAB-Vida (Portugal). Once confirmed that the different constructs were correct they were transferred to the pDONR207 vector using the BP clonase reaction (GATEWAY system, INVITROGEN) and transformed into the DH5α strain by the heat-shock method 13. The SSIV constructs were mobilized to the different final vector by the LR clonase reaction (GATEWAY system) and transformed into the DH5α strain by the heat-shock method. After confirmation of the plasmids by DNA restriction analysis, plasmids bearing the constructs with the CTP were transformed into the Agrobacterium tumefaciens strain C58 14 by electroporation. Constructs of SSIV without the CTP were transformed into the E. coli strain BL21 (DE3) ∆glgCAP, a derivative of the BL 21 (DE3) strain lacking the glgCAP operon, which encodes for the glycogen synthase, ADP-glucose pyrophosphorylase and glycogen phosphorylase enzymes 15. The final vectors employed with the SSIV polypeptides with the CTP were: pXNGW-(nYFP) 16, which allow the translational fusion of the N-terminal part of the fluorescent protein YFP (yellow fluorescent protein) to the C-terminal end of SSIV. pXCGW-(cCFP) 16, which allow the translational fusion of the C-terminal part of the fluorescent protein CFP (cyan fluorescent protein) to the C-terminal end of SSIV. And pEarleyGate 103 17, which allow the translational fusion of GFP (green fluorescent protein) to the C-terminal end of SSIV. In all cases, the stop codon of SSIV was eliminated to allow the translational fusion 15. The final vector for the SSIV constructs without the CTP was the pDEST17 vector (Thermo Fisher Scientific) vector, which allows the IPTG-inducible expression of the SSIV constructs and fuses a His5x tail at the N-terminal end of the protein.
Transient Expression in N. benthamiana- The saturated overnight bacterial cultures carrying the GFP or YFP/CFP construct moieties were each adjusted to a final OD600nm of 0.2 and then co-infiltrated with equal amounts of an Agrobacterium suspension carrying a p19 suppressor of post-transcriptional gene silencing, following the method of 16. The Agrobacterium suspensions were infiltrated into the leaves of 3- to 4-weekold N. benthamiana plants 17. The infiltrated plants were kept in a controlled growth chamber under the above conditions for 2 days until analysis by confocal microscopy.
Confocal microscopy- A DM6000 confocal laser scanning microscope (Leica Microsystems, http://www.leicamicrosystems.com) equipped with a 63X oil-immersion objective and a 20X objective was used to detect protein localization based on GFP fusion expression or protein-protein interaction in BiFC assays 18 performed in N. benthamiana mesophyll cells. GFP or YFP/CFP expression and chlorophyll autofluorescence imaging was performed by exciting the cells with an argon laser at 488 nm and detecting fluorescence emissions at 500–525 nm and 630–690 nm, respectively.
Immunoblot analysis- Proteins were transferred from an SDS-polyacrylamide gel to a polivinilidene fluoride (PVDF) membrane by electroblotting using a Trans-Blot Turbo transfer system (Bio-Rad, www.bio-rad.com) according to the manufacturer’s instructions. Membranes were incubated in blocking buffer at room temperature. The buffer contains 50 mL de TBS + Tween 20 0.1 % + 5% w/v nonfat dry milk. TBS composition: Tris-HCl 1M adjusted to pH 7.5 + NaCl 5M.
Blots were probed with anti-HIS5x antibody (Qiagen) (1:5000 dilution) 19 for 2 h at room temperature and washed three times for 15, 5 and 5 min respectively with TBS + Tween 20. Detection of hybridization was performed using Western Bright Quantum + peroxide (Advansta). Chemiluminescence was visualized using a Chemidoc XRS (Bio-Rad) apparatus running Quantity One software (Bio-Rad).
Protein expression in E. coli- Aliquots of overnight cultures (0.5 mL) were transferred to 50 mL Luria-Bertani (LB) medium containing 50 µg/mL kanamycin and 50 µg/mL ampicillin and only kanamycin for the negative control (strain not transformed). Cultures were incubated at 30°C until they reached an optical density of 0.5 at 600 nm. Cultures were induced to express SSIV polypeptides by adding 1 mM isopropyl-1-thiob-D-galactopyranoside (IPTG) at 30°C for 90 min. Bacterial cells from induced cultures were harvested by centrifugation, washed with 50 mM Hepes (pH 8.0), and resuspended in 100 mM Tricine buffer (pH 8.0), supplemented with 1mM phenylmethylsulfonyl fluoride (PMSF) and 5 µl/mL of protease inhibitor cocktail (Sigma-Aldrich). Cells were sonicated (BRANSON Digital Sonifier, 120 s, 10% intensity, 4°C) and then centrifuged for 30 min at 20,000 x g at 4°C. The supernatant constituted the crude extract.
Starch Synthase Activity- The glucosyltransferase activity of the SSIV constructs were assayed with two different substrates, amylopectin, and maltotriose. The reaction was performed in a final volume of 200 µl containing 100 mM Tricine (pH 8.0) 25 mM potassium acetate, 5 mM EDTA, 5 mM DTT, 0.5 mg/mL BSA, 0.5 M sodium citrate, 5 mM ADP-glucose and 10 mg/mL amylopectin, or 10 mg/mL maltotriose. The reaction was initiated by adding 10 µl of E. coli crude extracts obtained as described above. At 0 min (t0), 100 µl of the reaction was boiled for 10 min to stop the reaction. The rest of the assay (t10) was incubated at 30ºC for 10 min and the reaction stopped by boiling for 10 min. The ADP produced in the reaction was determined by HPLC using a Partisil 10 SAX column and a photodiode array detector.
RESULTS
As previously mentioned, we have analyzed the potential phosphorylation of the Tyr 515 and Ser 459 residues cited above and its possible role in the dimerization and SSIV activity. For this purpose, four different versions of SSIV were obtained changing the amino acid selected to an amino acid residue with a negative charge, to mimic the effect of phosphorylation, or to an amino acid without charge, to mimic the effect of the dephosphorylation of the residue. The substitutions carried out were: Tyr 515 changed to Phe (AtSSIV-Y515F), Tyr 515 changed to Glu (AtSSIV-Y515E). In the case of the Ser 459: Ser 459 changed to Ala (AtSSIV-S459A), and Ser 459 changed to Asp (AtSSIV-S459D). Polypeptides obtained were introduced into different vectors following the steps described in Experimental Procedures. The same versions were obtained without the chloroplast transit peptide to express them in E. coli. Such polypeptides were verified by sequencing and restriction enzyme. Due to the limited duration of this work, constructs with the different versions of Ser 459 were obtained but they were not studied in plant or E. coli.
Subcellular localization of the AtSSIV versions
The different versions of AtSSIV were cloned into the pEarleyGate 103 vector to fuse the marker protein GFP to the C-terminal end of each version. The plasmids obtained were transformed into the Agrobacterium tumefaciens strain C58, and these strains were used to agroinfiltrated Nicotiana benthamiana leaves. The expression pattern of the different versions fused to the GFP protein was analyzed by confocal microscopy. The N-terminal part of SSIV (AtSSIV-Nt), containing from Met 1 to Tyr 543 residues, was used as a positive control, as its expression pattern has been previously published and it is coincident with the expression pattern of the full-length SSIV protein 5. Figure 1, Panels A-C, shows the dot-like pattern of localization of AtSSIV-Nt. The same pattern was obtained for the AtSSIV-Y515F version of the protein (Panels D-F). However, no signal was obtained for the AtSSIV-Y515E version (data not shown).

Figure 1. Localization of different SSIV versions fused to GFP in Nicotiana benthamiana chloroplasts. Fluorescence images showing (left) GFP fluorescence (green), (middle) chlorophyll autofluorescence (red) and (right) merged images of proteins and chlorophyll in N. benthamiana leaves. The leaves were transiently transformed with a cDNA encoding AtSSIV-Nt (A-C), or AtSSIV-Y515F (D-E), fused to the GFP sequence.
Protein-protein interaction of the AtSSIV versions in Nicotiana benthamiana leaves.
The polypeptides AtSSIV-Y515F and AtSSIV-Y515E were cloned into the vectors pXNGW(-nYFP) and pXCGW(-cCFP). Plasmids obtained were transformed into the A. tumefaciens strain C58 and different combinations of the transformed strains were used to agroinfiltrate leaves as described in Experimental Procedures. The interaction between the different polypeptides tested was analyzed by confocal microscopy. As a positive control of protein-protein interaction, the plasmids pXNGW(-nYFP)_AtSSIV-Nt and pXCGW(- cCFP)_FBN1b (this plasmid contains the cDNA encoding for the fibrillins 1b gene fused to the C-terminal part of the marker gene CFP) were used. Figure 2, Panels A-C, shows the interaction between AtSSIV-Nt and the FBN1b protein. This interaction shows a dot-like pattern of localization, coincident with the pattern obtained for the AtSSIV-Nt polypeptide fused to the GFP protein. The AtSSIV-Y515F also showed interaction with FBN1b, with a similar pattern to that obtained for the AtSSIV-Nt polypeptide (Figure 2, Panels D-F). However, no interaction was observed between AtSSIV-Y515E and FBN1b polypeptides (Figure 2, Panels G-I).

Figure 2: Effect of mutation of Tyr 515 on the interaction of AtSSIV with the fibrillins 1b protein. Fluorescence images showing (left) YFP/CFP fluorescence (yellow); (middle) chlorophyll autofluorescence (red); and (right) merged images of YFP/CFP and chlorophyll fluorescence in N. benthamiana leaves. Leaves were co-infiltrated with Agrobacterium strains carrying a cDNA encoding the N-terminal part of the SSIV protein (A-C), or cDNA encoding the AtSSIV-Y515F protein (D-F), or cDNA encoding the AtSSIV-Y515E protein (G-I), fused to the N-terminal half of YFP, and the cDNA encoding the fibrillins 1b protein fused to the C-terminal half of CFP. The yellow signal indicates an interaction between the two polypeptides.
Our group has demonstrated that SSIV forms a dimer in vivo 12. Thus, we also analyzed if the mutation of Tyr 515 affected to the capability of SSIV to interact with another SSIV polypeptide. Each AtSSIV version, cloned into the pXNGW(-YFP) and pXCGW(-cCFP) vectors, were transformed into Agrobacterium strain C58 and co-infiltrated in Nicotiana leaves. We also analyzed if the two versions could interact with the unmodified version of the N-terminal part of SSIV, AtSSIV-Nt. Figure 3 shows that AtSSIV-Y515F was able to interact with the AtSSIV-Nt fragment (Panels A-C) and with another AtSSIV-Y515F polypeptide (Panels D-F). On the contrary, no interaction was observed between AtSSIV-Y515E and AtSSIV-Nt (Panels G-I) or AtSSIV-Y515E with another AtSSIV-Y515E polypeptide (Panels J-L).
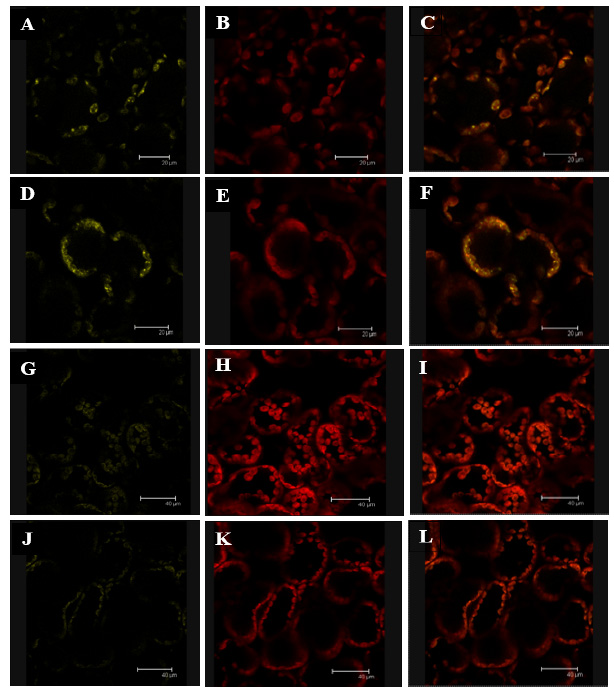
Figure 3: Effect of the mutation of Tyr 515 on the interaction AtSSIV-AtSSIV. Fluorescence images showing (left) YFP/CFP fluorescence (yellow); (middle) chlorophyll autofluorescence (red); and (right) merged images of YFP/CFP and chlorophyll fluorescence in N. benthamiana leaves. Leaves were co-infiltrated with Agrobacterium strains carrying the following combination of plasmids: Panels A-C: pXNGW(-nYFP)-AtSSIV-Nt and pXCGW(-cCFP)-AtSSIV-Y515F; Panels D-F: pXNGW(-nYFP)-AtSSIV-Y515F and pXCGW(-cCFP)-AtSSIV-Y515F; Panels G-I: pXNGW(-nYFP)-AtSSIV-Nt and pXCGW(-cCFP)-AtSSIV-Y515E; and Panels J-L: pXNGW(-nYFP)-AtSSIV-Y515E and pXCGW(-cCFP)-AtSSIV-Y515E. The yellow signal indicates an interaction between the two SSIV polypeptides.
Effect of Tyr 515 mutation on the SSIV activity.
To determine whether the mutation of Tyr515 could affect to the activity of AtSSIV, we cloned the two mutated versions into the pDEST17 vector, which allow the IPTG-inducible expression of the SSIV versions (AtSSIV-Y515F-WCTP, AtSSIV-Y515E-WCTP). In addition, this vector allows the tagging of the polypeptides expressed with a His5x tail tag at the N-terminal end of the proteins. The two AtSSIV versions were transformed into the E. coli strain BL21 (DE3) ΔglgCAP, which lacks the endogenous glycogen synthase activity. Selected clones containing the plasmids were cultivated and the expression of the AtSSIV versions were induced by IPTG. The levels of expression of the different versions were checked by immunoblot. One mL of each culture after the IPTG induction was collected, spun down in a microfuge and cells were resuspended in SDS-PAGE loading buffer and boiled for 10 min. The different samples were loaded into an SDS-PAGE gel; bands 45678were separated by electrophoresis and transferred to a PVDF membrane as described in Experimental Procedures. AtSSIV bands were located by hybridization with an antibody against the His5x tag. Results obtained are shown in Figure 5. The wild-type version of AtSSIV was also induced, although this cDNA was cloned into the pET45b (+) vector. The levels of expression of AtSSIV polypeptide was lower than levels of the mutated versions (Figure 5). Thus, to compensate for the different levels of expression in the enzymatic assay of the starch synthase activity of the different extracts, we quantified the intensity of the different bands using the Quantity-One software (BIO-RAD). This analysis indicated that the expression level of AtSSIV was 6-fold lower than the levels of the mutated versions. ( Figure 4)

Figure 4: Immunoblot analysis of AtSSIV-Y515F and AtSSIV-Y515E fragments expressed in E. coli. Cells were disrupted by sonication and centrifuged; the supernatant was analyzed on immunoblots probed with anti-His5x. Lanes: Mr: molecular weight marker. Ø: E. coli cells transformed with the empty expression vector pDEST17. A: cells transformed with the full-length AtSSIV cloned into the pET45b (+) vector, without the chloroplast transit peptide at time 0 and time 1h after addition of IPTG. B: crude extract of cells transformed with the AtSSIV-Y515F-WCTP polypeptide at time 0 and 1 h after adding IPTG. C: crude extract of cells transformed with the AtSSIV-Y515E-WCTP polypeptide fragment at time 0 and 1h of the IPTG induction.
The rest of the cells after the IPTG induction were collected and crude extracts were obtained as indicated in the Experimental Procedures to determine the starch synthase activity of the different AtSSIV polypeptides. Figure 5, shows the starch synthase activity obtained with the different AtSSIV polypeptides using amylopectin (Figure 5A) or maltotriose (Figure 5B) as substrate. It can be observed that the mutation of Tyr 515 drastically decreases the starch synthase activity, independently of the type of mutation. Using amylopectin as a primer, the positive control AtSSIV-WCTP protein displayed an activity of 16,24 ± 0,31 (SD) nmol.min-1, whereas the activity of the polypeptide AtSSIV-Y151F-WCTP was 0,35±0,02 nmol.min-1, and of the polypeptide AtSSIV-Y151E-WCTP was 0,36±0,01 nmol.min-1 (Figure 5 A). Using maltotriose as a primer, the control positive AtSSIV-WCTP protein had an activity of 29,12±1,93 (SD) nmol.min-1, whereas the activity of the polypeptide AtSSIV-Y151F-WCTP was 1,21±0,07 nmol.min-1, and of the polypeptide AtSSIV-Y151E-WCTP was 1,29±0,05 nmol.min-1 (Figure 5 B).

Figure 5: Starch synthase activity of the different AtSSIV polypeptides. Crude extracts of the same cultures analyzed in Figure 5 were used to determine the starch synthase activity using amylopectin (A) or maltotriose (B) as substrate. In order to compensate the different levels of expression of the polypeptides, 20 µl of crude extracts expressing the AtSSIV-Y515F-WCTP or the AtSSIV-Y515E-WCTP were used in the assay, whereas 120 µl was used in the case of crude extract expressing the wild-type version of AtSSIV. Values represent the mean±SD of three independent assays.
DISCUSSION
Initiation of the starch granule remains an uncharacterized process. Our group has proved that SSIV is involved in this process. The SSIV (AtSSIV) (At4g18240) of A. thaliana has a length of 1040 amino acids and an approximated molecular mass of 117.7 kDa 20. Previously this group has found a region localized between the long coiled-coils domains present at the N-terminal part of the protein and the C-terminal domain of SSIV 12. This region (CR) is highly conserved among the SSIV known to date. It extends from amino acid 471 to 515 of Arabidopsis SSIV and seems to be necessary for dimer formation. As outlined above, there is a tyrosine residue in the CR that is found in all the SSIV sequenced to date. This amino acid is at position 515 in the Arabidopsis SSIV amino acid sequence. In addition to the high degree of conservation, this amino acid displays a high probability to be phosphorylated. Another amino acid residue predicted to be phosphorylated is a residue of Ser at position 459. This amino acid is not in the CR but in the last of the four long coiled-coils domains observed in the N-terminal region of AtSSIV. This study aimed to analyze the effect of phosphorylation of these Tyr and Ser residues on the activity of the enzyme as well as on the pattern of localization of the enzyme and its capability to form dimers in vivo. To this purpose, we performed directed mutagenesis in which Tyr 515 and Ser 459 were changed to Phe and Glu in the case of the Tyr; and Ala and Asp in the case of the Ser residue. The charged residues aspartate and glutamate are used to mimic phosphorylation, whereas constitutive dephosphorylation is mimicked by Ala, Val, or Phe substitutions 21. Thus, we decided to change the Tyr residue by Glu in order to mimic the state of phosphorylation of the amino acid. To mimic the state of de-phosphorylation we paid attention to overall steric considerations and, to reduce the effect of unspecific effects, the Tyr residue was replaced by Phe, an amino acid with similar size to Tyr 22. Using the same principles, the Ser residue was changed to Asp and Ala, in order to mimic the states of phosphorylation and de-phosphorylation respectively (Figure 6). The analysis of the implication of the residues studied in the protein-protein interactions of AtSSIV was analyzed using the bi-molecular fluorescence complementation (BiFC) technology. A method that allows the possibility to assay protein-protein interactions directly in plant with comparative simplicity 23. The N-terminal part of SSIV (AtSSIV-Nt) was used as a positive control both in the BiFC assays and in the studies of localization using the GFP marker. Our group had demonstrated previously that this fragment displays the same pattern of localization (a dot-like pattern in the chloroplast) than the full-length AtSSIV protein and that interacts with the fibrillins 1 protein and with another AtSSIV-Nt polypeptide 10. The analysis of the localization of the AtSSIV-Y515F polypeptide fused to the GFP marker (Figure 1) indicates that the change of Tyr by Phe does not affect to the localization pattern of AtSSIV and suggests that this pattern would not depend on the phosphorylation of the protein. In the case of the mutated version where a Glu has changed the Tyr residue it is more difficult to obtain a conclusion, as no fluorescence signal was detected in this case. The integrity of the plasmid used was checked by sequencing, discarding a problem of translational fusion to the GFP protein or to a premature end of transcription as a consequence of a frameshift. Another possibility is that this mutated version of the AtSSIV polypeptide is unstable and undergoes proteolysis in the chloroplast. The two mutated versions of AtSSIV also displayed a distinct behavior concerning their capability to interact with the fibrillins 1b or to dimerize in vivo. Thus, the AtSSIV-Y515F polypeptide was able to interact with FBN1b (Figure 2) as well as with the N-terminal part of the wild-type version of AtSSIV and with another AtSSIV-Y515F polypeptide (Figure 3).

Figure 6. Scheme showing the amino acid changes used in the directed mutagenesis of Arabidopsis thaliana SSIV. Purple box corresponds to coiled-coil region (with Ser 459 amino acid marked in blue). Orange box indicates the highly conserved region (CR) found in SSIV proteins (with Tyr 515 amino acid marked blue). Part of the amino acid and nucleotide sequence of region depicted are shown at the bottom of the picture, together with the point mutation introduced to obtain the different mutated version of AtSSIV.
On the contrary, no protein-protein interaction was observed in the case of the AtSSIV-Y515E polypeptide (Figures 2 and 3). These results could indicate that the phosphorylation of the Tyr 515 would prevent the formation of a dimer and hence the interaction with FBN1b. However, we cannot discard that, as suggested by the localization studies, the substitution of the Tyr residue by a Glu makes unstable the protein and this is degraded in the chloroplast so that no interaction with any other polypeptide can be detected.
The analysis of the enzymatic activity of the mutated versions in E. coli shows that the change of the Tyr 515 residue by either Phe or Glu drastically decreases the activity of AtSSIV (Figure 5). In this study, both polypeptides were expressed and stably accumulated in E. coli, as it can be observed in Figure 5. However, E. coli and the chloroplasts are considerably distinct environments, and the stability of the AtSSIV-Y515E polypeptide can be different in the bacteria and in the chloroplast. Our group has demonstrated that the elimination of the CR prevents the formation of a dimer and decrease the enzymatic activity of AtSSIV drastically, suggesting that is the dimer the form enzymatically active 12. Our results suggest that the change of the Tyr 515 residue by a Phe does not affect the formation of a dimer but reduces the activity of the polypeptide considerably. A similar decrease was observed when the amino acid was changed to a Glu residue, indicating that this residue is necessary for the AtSSIV activity rather than for the formation of a dimer. Our group has shown that AtSSIV displays more affinity for the maltooligosaccharide maltotriose than for amylopectin 10, and for that reason, the activity of the mutated versions was analyzed with both substrates. Our results indicate that the mutation of Tyr 515 affected negatively to the activity of AtSSIV irrespectively of the substrate employed.
CONCLUSIONS
The Tyr 515 residue seems to be necessary for the starch synthase activity of AtSSIV and further studies are necessary to elucidate the effect of the change of this Tyr by a Glu in the plant. For instance, the stability of this mutated version in-plant could be followed by immunoblot using an antibody against the His5x tag or the AtSSIV protein available in our group.
REFERENCES
1. Zeeman SC, Kossmann J and Smith AM. Starch: its metabolism, evolution, and biotechnological modification in plants. Annu Rev Plant Biol 2010; 61: 209-234. DOI: 10.1146/annurev-arplant-042809-112301.
2. Ball S, Colleoni C, Cenci U, et al. The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J Exp Bot 2011; 62: 1775-1801. DOI: 10.1093/jxb/erq411.
3. Ball SG and Morell MK. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 2003; 54: 207-233. DOI: 10.1146/annurev.arplant.54.031902.134927.
4. D'Hulst C and Merida A. The priming of storage glucan synthesis from bacteria to plants: current knowledge and new developments. New Phytol 2010; 188: 13-21. DOI: 10.1111/j.1469-8137.2010.03361.x.
5. Gamez-Arjona FM, Raynaud S, Ragel P, et al. Starch synthase 4 is located in the thylakoid membrane and interacts with plastoglobule-associated proteins in Arabidopsis. Plant J 2014; 80: 305-316. DOI: 10.1111/tpj.12633.
6. Hirose T and Terao T. A comprehensive expression analysis of the starch synthase gene family in rice (Oryza sativa L.). Planta 2004; 220: 9-16. DOI: 10.1007/s00425-004-1314-6.
7. Leterrier M, Holappa LD, Broglie KE, et al. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: functional and evolutionary implications. BMC Plant Biol 2008; 8: 98. DOI: 10.1186/1471-2229-8-98.
8. Edwards A, Marshall J, Sidebottom C, et al. Biochemical and molecular characterization of a novel starch synthase from potato tubers. The Plant journal 1995; 8: 283-294.
9. D'Hulst C and Merida A. Once upon a prime: inception of the understanding of starch initiation in plants The synthesis and breakdown of starch SEB Essential Reviews in Experimental Biology 2012; 1: 55-76.
10. Szydlowski N, Ragel P, Raynaud S, et al. Starch granule initiation in Arabidopsis requires the presence of either class IV or class III starch synthases. Plant Cell 2009; 21: 2443-2457. DOI: 10.1105/tpc.109.066522.
11. Youssef A, Laizet Y, Block MA, et al. Plant lipid-associated fibrillin proteins condition jasmonate production under photosynthetic stress. Plant J 2010; 61: 436-445. DOI: 10.1111/j.1365-313X.2009.04067.x.
12. Raynaud S, Ragel P, Rojas T, et al. The N-terminal Part of Arabidopsis thaliana Starch Synthase 4 Determines the Localization and Activity of the Enzyme. J Biol Chem 2016; 291: 10759-10771. DOI: 10.1074/jbc.M115.698332.
13. Froger A and Hall JE. Transformation of plasmid DNA into E. coli using the heat shock method. J Vis Exp 2007: 253. DOI: 10.3791/253.
14. Morán-Zorzano MT, Alonso-Casajus N, Munoz FJ, et al. Occurrence of more than one important source of ADPglucose linked to glycogen biosynthesis in Escherichia coli and Salmonella. FEBS Lett 2007; 581: 4423-4429. DOI: 10.1016/j.febslet.2007.08.017.
15. Earley KW, Haag JR, Pontes O, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 2006; 45: 616-629. DOI: 10.1111/j.1365-313X.2005.02617.x.
16. Silhavy D, Molnár A, Lucioli A, et al. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. . EMBO J 2002; 21: 3070-3080.
17. Marillonnet S, Thoeringer C, Kandzia R, et al. Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat Biotechnol 2005; 23: 718-723. DOI: 10.1038/nbt1094.
18. Yuan L, Gu R, Xuan Y, et al. Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell 2013; 25: 974-984. DOI: 10.1105/tpc.112.108027.
19. Roldan I, Wattebled F, Mercedes Lucas M, et al. The phenotype of soluble starch synthase IV defective mutants of Arabidopsis thaliana suggests a novel function of elongation enzymes in the control of starch granule formation. Plant J 2007; 49: 492-504. DOI: 10.1111/j.1365-313X.2006.02968.x.
20. Rose A, Manikantan S, Schraegle SJ, et al. Genome-wide identification of Arabidopsis coiled-coil proteins and establishment of the ARABI-COIL database. Plant Physiol 2004; 134: 927-939. DOI: 10.1104/pp.103.035626.
21. Dissmeyer N and Schnittger A. Use of phospho-site substitutions to analyze the biological relevance of phosphorylation events in regulatory networks. Methods Mol Biol 2011; 779: 93-138. DOI: 10.1007/978-1-61779-264-9_6.
22. Murray R, Bender D, Botham K, et al. Harper´s Illustrated Biochemistry 28 ed.: McGraw-Hill, 2009.
23. Schutze K, Harter K and Chaban C. Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 2009; 479: 189-202. DOI: 10.1007/978-1-59745-289-2_12.
Cristina Calderón1, Ángel Mérida2.
1. Escuela Superior Politécnica de Chimborazo. Panamericana Km 1 ½. Riobamba. Ecuador. E-Mail: [email protected]. ORCID: https://orcid.org/0000-0002-8574-103X
2. Instituto de Bioquímica Vegetal y Fotosíntesis. Consejo Superior de Investigaciones Científicas University of Sevilla. Avda. Américo Vespucio, 49. 41092-Sevilla. Spain. E-Mail: [email protected]
