2021.06.04.18
Files > Volume 6 > Vol 6 No 4 2021
Phylogenetic and molecular evolutionary analysis of SENV DNA isolated from Iraqi Hepatitis Patients
Arwa Mujahid Al-Shuwaikh 1, Ealaf Abbas khudair 1, Dalya Basil Hanna 2.
Available from: http://dx.doi.org/10.21931/RB/2021.06.04.18
ABSTRACT
SEN Virus (SENV) is a newly discovered group of transmissible, hepatotropic, single-stranded, circular, non-enveloped DNA viruses that are distantly linked to the widely distributed Torque Teno Virus (TTV) family. This research aimed to use nucleotide sequencing to identify the genetic alterations of SEN-V and to investigate the similarities between isolates. Seven DNA samples of SENV, which were previously extracted from blood of post transfusion hepatitis, were used to identify the genetic variation of SEN-V by nucleotide sequencing. According to the current analysis results, specific primer pairs were used to detect SENV DNA sequences isolated from Iraqi patients with hepatitis; however, those specific primers can also detect two new variants of SENV that are closely related to the Torque Teno Virus. In addition, four SENV isolates showed several substitution mutations, and one of them revealed the replacement of Proline (P) at position 11 with Serine (S). Only one local isolate of SENV was 100% identical to the Iranian isolate (GenBank acc. no. GQ452051.1) from thalassemia.
Key words. SEN-V, Sequencing, Phylogenetic tree.
INTRODUCTION
In addition to well-known A to E hepatitis viruses, there is growing evidence that newer hepatitis viruses may exist and have a role in this disease1. The SEN virus (SENV) is a member of a newly discovered family of hepatotropic, non-enveloped, single-stranded (ss) DNA of negative polarity viruses that was reported as non-A to E hepatitis virus in 19992. SEN-V is a circular 3,900 nucleotides, possibly belonging to the Anelloviridae family, including Torque Teno virus (TTV), TTV-like minivirus, SANBAN virus and SAN YONBAN virus. Among nine detected genotypes of SEN-V; genotypes D and H are more frequent in hepatitis cases3,4,5. SENV and TTV have related genomic structures; SENV's nucleotide sequence found two dominant, partially overlapping open reading frames (ORF1 and ORF2)6,7,8,9.
About 20% of hepatic infections are not associated with hepatitis viruses (A–E), which may be interpreted as other viruses' involvement. Wide ranges of SENV infections are reported in individuals with liver disease. SENV is ubiquitous with distinct geographic variations. Epidemiological studies have evidenced wide ranges of SENV in other pathological conditions, such as autoimmune diseases, respiratory conditions, cancer, thalassemia patients and patients on maintenance hemodialysis. In spite of several studies, the pathogenetic role of SENV has not yet been clarified10,11,12,13. The current study aims to detect the genetic changes in SEN-V and/or similarities between isolates in hepatitis patients.
METHODS
Selection of samples
Seven (7) DNA samples of SENV, which were previously extracted from the blood of post-transfusion hepatitis, those DNA samples were used to identify the genetic alterations of SENV by Sanger nucleotide sequencing. Until used, these specimens were held in a deep freeze (-44°C).
Detection of SENV DNA
The samples were taken out of the freezer and thawed. For the isolation and purification of DNA from specimens, the Viral Nucleic Acid Extraction Kit II (Cat. #VR00, Geneaid, Taiwan) was used. The operation was carried out following the manufacturer's instructions. According to Karimi-Rastehkenari et al.2, SENV DNA was determined using nested polymerase chain reaction (PCR) with SENV-specific primers. For amplification, PCR fragment was chosen, which was expected to cover 119 bp of the SENV. SENV was amplified for the first round with forwarding primer AI-1F (5’-TWCYCM AAC GACCAG CTA GAC CT-3'; W = A or T, Y = C or T, M = A or C) and reverse primer AI-1R (5’-GTT TGT GGTGAG CAG AAC GGA-3'), while for the second-round PCR amplification with specific forward and reverse primers H-1020F (5’-TTT GGC TGC ACC TTC TGG TT-3') and H-1138R (5’-AGA AAT GAT GGG TGA GTG TTA GGG-3'), respectively.
Genomic amplification
According to Khudair et al.11, DNA amplification reactions were carried out using nested traditional PCR. For the first and second round reactions, the cycling conditions were similar as follows: initial denaturation at 95°C for 5 minutes for 1 cycle, followed by of DNA amplification by sequentially heating for denaturation of DNA template at 95°C for 30 seconds, annealing at 60°C for 45 seconds, and extension at 72°C for 45 seconds for 35 cycles, then final extension 72˚C for 5 minutes for 1 cycle. The amplicons were electrophoresed in a 1 percent agarose gel for visualization. The band size was determined using an ultraviolet (UV) transilluminator and a direct comparison to a 100 bp DNA marker. One band of 119 bp DNA was found in SENV DNA positive samples, as shown in figure (1). The amplified SENV DNA samples have a code number of (1, 5, 68, 100, 122, 123, and 136). These seven samples were sent to Macrogen Company in South Korea and their corresponding primers for sequencing.
Analysis of sequences
The nucleotide sequence similarities between isolates and the documented reference sequence were determined in this study using the BLAST program (http://www.ncbi.nih.gov) and the BioEdit program. Nucleotide sequences were translated to a protein sequence using the online Expasy translate server. Using the NCBI-BLAST server, a specific comprehensive viral tree was built, in which the observed viral variant was compared to their neighbor homologous viral sequences14. Then, using Clustal Omega for making multiple sequence alignments. In addition, FigTree (http://tree.bio.ed.ac.uk/software/figtree/) a program was used to graphically view phylogenetic trees, including the observed variant, as a polar cladogram. In the comprehensive tree, the viral sequences of each classified phylogenetic species group were colored accordingly.
RESULTS
Sequencing of SENV local isolates
According to the sequencing company's instructions, seven SENV isolates' 119-bp PCR second round amplicons were commercially sequenced from both ends. Only specific chromatographs from ABI sequence files were studied further, ensuring that the annotation and differences were not due to PCR or sequencing errors. The exact location and other aspects of the retrieved PCR fragment were established by comparing the observed DNA sequences of both local specimens with the retrieved DNA sequences of SENV (GenBank acc.GQ452051.1).
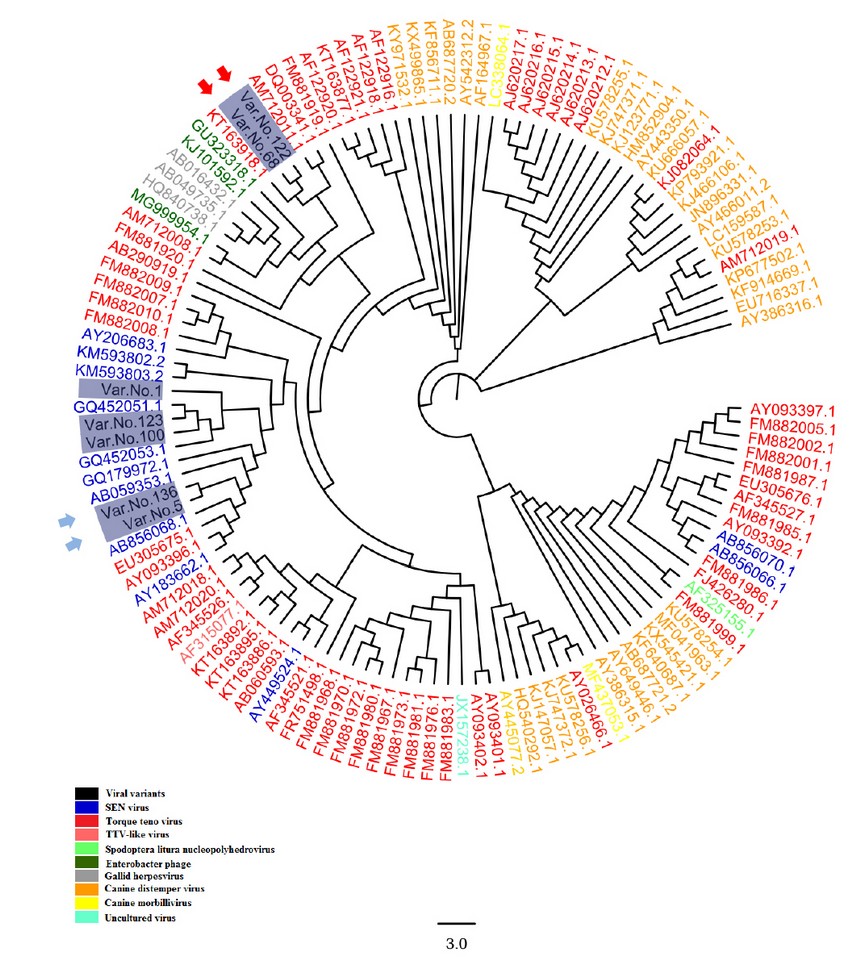
The SENV sequences were highlighted in table (1); the grey-colored regions correspond to forward and reverse primers, respectively. The forward primer is positioned in the positive strand, while the position of the reverse primer was in the negative counterpart. The alignment results of all sequenced samples revealed the presence of seven mutations, in which substitution was observed, as shown in table (2). The sequencing chromatogram of the observed point mutation was documented in these amplicons, as shown in figure (2). The exact positions of observed mutation were listed in the NCBI reference sequences figure (3) to summarize all of the results obtained from the sequenced 119 bp fragment. Interestingly, The NCBI Blast server disclosed that there were noticeable differences between the previously sequenced samples (Var. Nos. 1, 5, 100, 123, and 136) and the (Var. No. 68 and 122), as shown in figure (4).
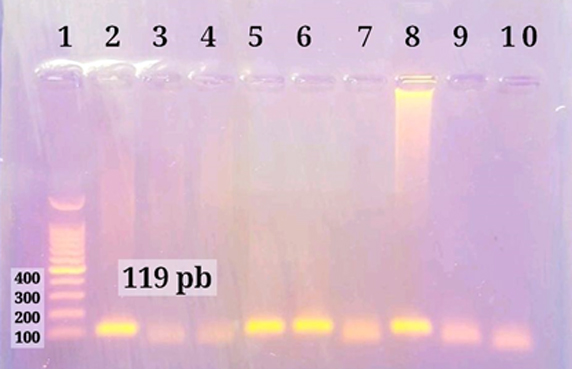
Figure 1. Agarose gel electrophoresis of PCR products second round. Line 1: 100 bp ladder. Lines 2,5,6 and 8 SENV-H positive 119 bp. Lines 3,4,7,9 and 10 negative samples.

Table 1.The position and length of the PCR amplicon used to amplify SENV. The amplified sequence was extended from 1 into 119 of the NCBI reference DNA sequence (GenBank acc. no. GQ452051.1). The grey-colored regions refer to forward and reverse primers, respectively.
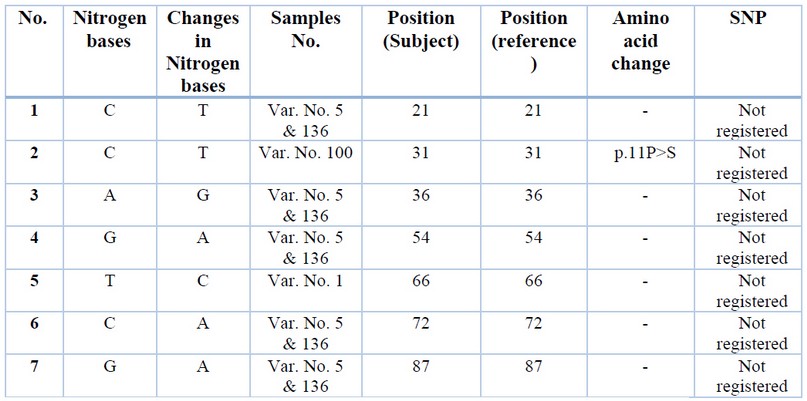
Table 2. The observed point mutations compared to the NCBI referring sequences of the SENV (GenBank acc. no.GQ452051.1).
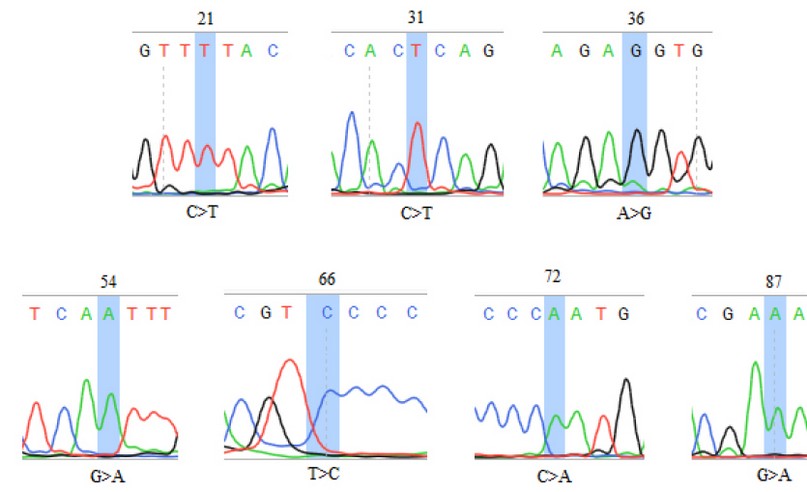
Figure 2. The DNA chromatogram sequence of local isolates of SENV. The locations of the observed substitution mutations were highlighted. The symbol ">" denotes a substitution mutation.

Figure 3. The DNA sequences of the observed local viral isolates were aligned with the corresponding reference sequences of the SENV. The substitution mutations were highlighted in the PCR products according to their positions. The NCBI referring sequence (GenBank acc. no.GQ452051.1) is denoted by the symbol "ref.seq." The colored arrows indicate substitution mutations that have been found.
DISCUSSION
Viral hepatitis is a worldwide disease that presents a severe threat to human health. To date, five hepatitis viruses have been discovered, ranging from A to E, accounting for 80% to 90% of hepatitis cases. However, none of the hepatitis viruses were found in the remaining 10% to 20% of cases with classic viral hepatitis symptoms, indicating that other viruses, such as SENV, are likely to play a role in the pathogenesis of non-A-E hepatitis 3,13.
In this study, the total number of aligned nucleic acid sequences in the comprehensive phylogenetic tree figure (4), regardless of viral variants, was 123 sequences including seven different local SENV isolates from viral hepatitis patients. in addition to both SENV and TTV sequences, this viral-specific phylogenetic tree consists of several viral species, namely TTV-like viruses, Spodoptera Litura Nucleopolyhedroviruses, enterobacter phages, Gallid herpes viruses, Canine distemper viruses, as well as uncultured viruses 15,16. The comprehensive phylogenetic tree was found that observed viral variants encompassed two-variable phylogenetic positions; the first one is located in the SENV referring sequencing group, while the other group is located in the TTV referring sequencing counterpart. The current study-based phylogenetic tree indicated that two main categories are originated from the presence of specific differences between the sequences of both viral groups.
The first viral group is just derivatives of the main SENV referring sequences. This sort of difference belongs to the observed DNA variations that were revealed in five sequenced viral samples. The Var. No. 1 was exerted only one variation from the SENV referring sequence (g.66T>C). In addition, the situation is the same in Var. No.100 that showed transition replacement of pyrimidine base with another pyrimidine (g.31C>T) which led to replacement of Proline (P) at position 11 with Serine (S). Whereas Var. Nos. 5 and 136 have exerted more variations (g.21C>T, g.36A>G, g.54G>A, g.72C>A, and g.87G>A), as shown in table (2). Thus, both Var. No. 5 and Var. No. 136 were occupied a distinctive position in the tree. Such position was however located within SENV phylogenetic regions, as shown in figure (4).
The both local Var. Nos. 68 and 122 were distant from the SENV phylogenetic position. In contrast they showed similarity to Torque Teno virus (acc. no. TK163918.1). The current finding was that two samples No. 68 and 122 deviated from the expected SENV sequenced samples into the unexpected Torque Teno viruses. This is perhaps due to several reasons; SEN and TTV have many in common sequences as they share many homologies. The genomic sequences of SENV are relatively similar to TTV, and both are classified within the Anelloviridae family3. Add to that, both SENV and TTV are single-stranded non-enveloped DNA viruses of 3,900 nucleotides 2,17,18,19,20. The extraordinary diversity data obtained from the present study follows other reports that notified such a particular interconnection between SENV and TTV 7,21. In addition, The high genomic homology seen between these two SENV isolates and several TTV isolates could point to SENV's evolutionary history concerning TTV1. According to one study, some of the TTV-related isolates can be pathogenic15. Through specifically designed primers pairs were utilized in this study to detect SENV DNA sequences, it's probably that such specific primers may detect the closely related TTV sequences instead22.
CONCLUSIONS
This study showed that several silent point mutations had been found in SENV ORF1 region. However, only one mutation in isolate No.100 led to the replacement of Proline with Serine. The present phylogenetic tree provided a clue about the identity of these local isolates, and a more extensive scale screening study is required for more information about the pattern of relatedness between SENV sequences and their TTV counterparts in different diseases in our region.
Acknowledgments and funding
The authors acknowledge the technical assistance of the Microbiology Department, College of Medicine, Al Nahrain University and the Department of Clinical Laboratory Sciences, College of Pharmacy, Mustansiriyah University. L'Oreal-UNESCO supported this work for the Women in Science program.
Conflict of Interest
There are no potential conflicts of interest for the writers to disclose.
REFERENCES
1. Khudair EA, Al-Shuwaikh AM. Detection of SEN virus (SEN-V) and Torque Teno virus (TTV) Co-Infection and Liver Enzyme in a Sample of Hepatitis Patients. Al Mustansiriyah Journal of Pharmaceutical Sciences. 2020; 20(3):28-35.
2. Karimi-Rastehkenari A, Bouzari, M. High frequency of SEN virus infection in thalassemic patients and healthy blood donors in Iran. Virology Journal. 2010; 7(1):1-7.
3. Hosseini SA, Bouzari M. Detection of SENV virus in healthy, hepatitis B-and hepatitis C-infected individuals in Yazd Province, Iran. Iranian biomedical journal. 2016; 20(3):168.
4. Sagir A, Kirschberg O, Heintges T, et al. SEN virus infection. Reviews in medical virology. 2004;14(3):141-148.
5. Tanaka Y, Primi D, Wang RY, et al. Genomic and molecular evolutionary analysis of a newly identified infectious agent (SEN virus) and its relationship to the TT virus family. The Journal of infectious diseases. 2001 1;183(3):359-67.
6. Kojima H, Kaita KD, Zhang M, et al. Genomic analysis of a recently identified virus (SEN virus) and genotypes D and H by polymerase chain reaction. Antiviral research. 2003; 60(1):27-33.
7. Diniz-Mendes L, Devalle S, Niel C. Genomic characterization of a Brazilian TT virus isolate closely related to SEN Virus-F. Memórias do instituto oswaldo cruz. 2004; 99(3):301-306.
8. Xu D, Tian DY, Huang YC. Phylogenetic Analysis of Different Open Reading Frame of SEN Virus. Chinese Journal of Integrated Traditional and Western Medicine on Liver Diseases. 2005; (4):8.
9. Basheer SH, Elzaki SG, Abaker NM, et al. Prevalence of SEN virus infection among volunteer blood donors in Nyala teaching hospital, Southern Darfur state, Sudan. Scholars Journal of Applied Medical Sciences. 2017;5:4441-5.
10. Bourinbaiar AS, Jirathitikal V, inventors; Immunitor USA Inc, assignee. Immunotherapy and prevention of autoimmune hepatitis. United States patent application US 12/662,663. 2010; 9.
11. Khudair EA, Al-Shuwaikh AM, Farhan NM. Detection and Genotyping of SEN Virus among Patients with Hepatitis and Healthy Blood Donors from Baghdad, Iraq. Jordan Journal of Biological Sciences. 2019; 12(3).
12. dos Santos Bezerra R, Santos EV, Silveira RM, et al. Molecular prevalence and genotypes of human pegivirus-1 (HPgV-1) and SENV-like viruses among multiply transfused patients with beta-thalassemia. Transfusion and Apheresis Science. 2019; 13:102697.
13. El-Sayed HA, ElHamid NA, ElBadrawy EM, et al. Prevalence and clinical significance of SEN-virus and TT-virus infection in chronic HCV patients. The International Arabic Journal of Antimicrobial Agents. 2019; 9(3).
14. Ye J, McGinnis S, Madden TL. BLAST: improvements for better sequence analysis. Nucleic acids research. 2006; 34(suppl_2):W6-9.
15. Tanaka Y, Primi D,Wang RYH, Umemura T, Yeo AET, Mizokami M, Alter HJ, Shih JW-K. Genomic and Molecular Evolutionary Analysis of a Newly Identified Infectious Agent (SEN Virus) and Its Relationship to the TT Virus Family. The Journal of Infectious Diseases. 2001;183:359–367.
16. He W, Gao Y, Wen Y, Ke X, Ou Z, Li Y, He H, Chen Q. Detection of Virus-Related Sequences Associated With Potential Etiologies of Hepatitis in Liver Tissue Samples From Rats, Mice, Shrews, and Bats. Frontiers in Microbiology. 2021; 12:653873.
17. AbuOdeh R, Al-Mawlawi N, Al-Qahtani AA, et al. Detection and genotyping of torque teno virus (TTV) in healthy blood donors and patients infected with HBV or HCV in Qatar. Journal of medical virology. 2015;87(7):1184-1191.
18. Khudair EA, Al-Shuwaikh AM, Farhan NM. Detection of TTV antigen in patients with hepatitis HBV and HCV. Iraqi Journal of Medical Sciences.2019;17(1):43-49.
19. Zheng MY, Lin Y, Li DJ, Ruan HB, et al. TTV and HPV co-infection in cervical smears of patients with cervical lesions in littoral of Zhejiang province. Chinese journal of experimental and clinical virology. 2010; 24(2):110-112.
20. Pirouzi A, Bahmani M, Feizabadi MM, Afkari R. Molecular characterization of Torque teno virus and SEN virus co-infection with HIV in patients from Southern Iran. Revista da sociedade brasileira de medicina tropical. 2014;47(3):275-279.
21. Hino S, Miyata H. Torque teno virus (TTV): current status. Reviews in medical virology. 2007;17(1):45-57.
22. Bouzari M, Baygloo SN. Primers designed for detection of TT virus also detect SEN virus. Research Journal of Biological Sciences.2008; 3:1063-1066.
Received: 14 June 2021
Accepted: 5 August 2021
Arwa Mujahid Al-Shuwaikh 1, Ealaf Abbas khudair 1, Dalya Basil Hanna 2.
1Microbiology Department, College of Medicine, Al-Nahrain University, Baghdad – Iraq.
2Department of Clinical Laboratory Sciences, College of Pharmacy, Mustansiriyah University, Baghdad, Iraq.
Correspondence to Assist. Prof. Dr. Arwa Mujahid Al-Shuwaikh.
https://orcid.org/0000-0002-2468-9629
