2023.08.03.113
Files > Volume 8 > Vol 8 No 3 2023
Using recombinant DNA technology in bacterial identification from vaginal swabs
1 Nineveh Health Department / Mosul / Iraq; Email: [email protected].
2 Department of Biology, College of Science, University of Mosul/ Mosul /Iraq
* Correspondence: [email protected] Tel.: +9647736834730
Available from: http://dx.doi.org/10.21931/RB/2023.08.03.113
ABSTRACT
Many bacterial pathogens fail to be isolated on cultural media for several reasons. Accordingly, specimens are misdiagnosed as unfavorable, where they might carry some pathogenic microbes. Developing new strategies for bacterial identification is essential to provide better health quality. Therefore, this study aimed to employ Thymine-Adenine cloning (TA cloning) in bacterial diagnosis from vaginal swabs. For this purpose, 20 vaginal swabs were collected from patients hospitalized in Mosul city hospitals. Swabs were exposed to traditional methods for bacterial isolation and identification. Results showed that 55% of the samples produced bacterial colonies on cultural media, while 45% failed to produce colonies.
On the other hand, genomic DNA was directly isolated from the identical 20 vaginal swab specimens and was used to amplify the 16S rRNA gene. PCR products were cloned to pGEM-T easy vector via TA cloning and sequenced to identify the bacterial pathogens involved. Results showed that all vaginal swabs amplified 16S rRNA gene products, and when cloned to pGEM-T easy vector and sequenced, multiple miss-diagnosis cases were noticed. This method requires more time and skills and gives highly accurate results that traditional forms of isolation and identification do not provide.
Keywords: 16S rRNA, bacterial identification, vaginal swab.
INTRODUCTION
Health care and control of infection transmission rely on the correct methods in identifying the causative agent. Culture and biochemical tests are currently the gold standards for bacterial pathogen detection in diagnostic microbiology labs. This approach is commonly used in hospitals in most middle-income nations1. Early detection of pathogens in clinical samples is necessary for improved diagnosis and management of diseases2. Due to a variety of reasons, some organisms may need help to detect using standard culture techniques employed in most laboratories. Some significant reasons are strict growth requirements, non-viable organisms, the inhibition of pathogenic organisms caused by the production of bacteriocins by other microbes or the administration of antibiotics by the patient3. Traditional techniques for detecting bacterial pathogens are time-consuming and frequently require an ideal bacterial concentration of at least 105 cfu/ml in the sample to generate accurate results; otherwise, false adverse effects are expected4. In research that expanded for 27 years, researchers found that when traditional identification methods were used in identifying bacterial meningitis, around 13% of the time, identification may fail 5. The possibility of acquiring false negative results increases with the decrease of bacterial concentration present in the specimen. Regardless of the pathogen present in CSF samples, only 25% were detected correctly when the concentration decreased below 103 cfu/ml6.
The last ten years of the twentieth century they were allowed for an exponential increase in the knowledge of molecular biology techniques following the cellular and protein era of the 1970s and 1980s. This explosion of technologies has had major consequences and has allowed for significant developments in many areas of life sciences, including bacteriology7. Introducing PCR tests in clinical laboratories was an important step that helped directly detect bacteria from clinical specimens and find genes encoding antibiotic resistance8. A recent publication using direct PCR on CSF and blood samples could identify pathogens not previously detected using traditional microbiological techniques9. However, CSF and blood specimens are sterile; once infected, the infection is usually caused by a single pathogen. Unlike these samples, vaginal swabs contain a consortium of microbes, thereby direct PCR on these samples will not be an appropriate idea. One way to sequence 16S rRNA gene fragments is by sequencing next-generation sequencing. High throughput technology and bioinformatics identify pathogens accurately and may identify the presence of novel uncultured pathogens in the sample.
Nevertheless, this technique is costly, requires high knowledge in analyzing genome sequences and is time-consuming as well10. This paper introduced a new method to separate mixed sequences of 16S rRNA fragments obtained from direct amplification of 16S rRNA gene from vaginal swabs. This method is based on cloning DNA fragments on a pGEM-T easy vector via TA cloning followed by DNA sequence.
MATERIALS AND METHODS
Collection of clinical specimens
A total of 20 vaginal swabs were collected from different clinical cases of hospitalized patients from Mosul-Iraq from December 2021 to April 2022. All specimens were collected in appropriate sterile swabs containing 5 mL of normal saline. Swabs were transported to the laboratory in cooled containers for bacterial identification using traditional and molecular methods3.
Traditional culture methods for bacterial isolation
Samples from vaginal swabs were streaked on three different media (blood agar, chocolate agar, and MacConkey agar). Plates were incubated at 37°C for 24–48 hours. Traditional bacterial colony diagnoses depended on morphological features and simple biochemical tests, including oxidase and catalase. To examine the morphology of the cells and how they interacted with the dye, pure cultures were Gram-stained11.
Molecular techniques
DNA Extraction
Genomic DNA was isolated from vaginal swabs using a Geneaid isolation kit provided by (Geneaid company, Taiwan). Genomic DNA was directly isolated from vaginal swab samples by taking 1mL of the normal saline containing the soaked swab; then, genomic DNA isolation was conducted as recommended by the manufacturer. Genomic DNA was kept at -20°C until further used12.
Polymerase Chain Reaction
PCR was conducted in a 20µl volume reaction using GoTaq G2 Green Master Mix supplied by Promega (USA) to amplify the 1495bp region of the 16S rRNA gene. The universal primers used were 27F AGAGTTTGATCMTGGCTCAG and 1522R AAGGAGGTGATCCARCCGCA. The final concentration of primers and the total amount of template DNA were added as recommended by the manufacturer. The PCR program used to amplify the 16S rRNA gene was set as follows: initial denaturation at 95oC for 3 minutes followed by 30 cycles of amplification, including a denaturation step at 95oC for 30 seconds, annealing at 55oC for 30 seconds, and extension at 72oC for 1 minute. A final extension step was set at 72oC 9. PCR products were separated on 1% agarose gel and stained with Midori Green Advance DNA stain. A 100bp DNA marker (New England Biolabs) was used as a molecular weight marker13.
Thymine: Adenine cloning (TA Cloning)
The 16S rRNA gene fragment was cloned via the TA cloning kit supplied by Promega (USA) to the linearized plasmid, pGEM-T easy vector (3Kb). The reaction was conducted in a 10µl volume sample containing 5µl 2X rapid ligation buffer (T4 DNA Ligase), 1µl pGEM-T easy vector (50ng), 1µl T4 DNA ligase, a variable amount of insert DNA required to create a 3:1 insert: vector ratio, and the final volume was completed to 10 µl with nuclease-free distilled water. The ligation mixture was incubated overnight at 4oC as recommended by Promega14. A 3 µL sample from the ligation mixture was used to transform E. coli DH5α competent cells and plated on LB agar plates containing ampicillin (50µg/ml) and Xgal (80µg/ml) as mentioned elsewhere15.
Plasmid isolation
Plasmid DNA was isolated from three colonies of white E. coli DH5α that appeared on LB agar plates containing ampicillin and X-gal using the plasmid DNA isolation kit provided by Promega (USA) following the recommended steps of the company. Plasmid DNA was digested with EcoRI to identify the presence of the 16 rRNA gene fragment insert. Verified clones were stored at -20°C until further used 15.
DNA sequencing and Homology search
pGEM-T easy vector clones containing the 16S rRNA gene fragment were purified and sent for sequencing at Psomagene sequencing company (USA) using the primers T7 promoter 5’ TAATACGACTCACTATAGGG 3’ and SP6 5’ ATTTAGGTGACACTATAG 3’. Retrieved sequences were searched for homology against published genes submitted in GenBank using the BLAST tool at NCBI.
RESULTS AND DISCUSSION
Traditional methods for isolation
Traditional bacterial isolation and identification methods from vaginal swabs detected 11 out of 20 positive samples, as shown in Table 1. Lactobacillus was found to be the significant member in vaginal swab cultures as it was detected in 9 out of the 11 positive cultures using traditional methods for isolation, which is acceptable as this genus, along with Bifidobacterium which is the most prevalent normal flora found in the vagina and the presence of lactobacilli helps in eliminating other pathogens16. Recent studies have shown that the administration of Lacticaseibacillus rhamnosus and Lacticaseibacillus paracasei probiotics increased the number of lactobacilli and decreased the numbers of some vaginal pathogens, particularly enterococci, staphylococci, Gardnerella and Candida17.
The pathogens detected in vaginal swabs using traditional methods were Staphylococcus, Enterococcus and E. coli. These pathogens are well known for causing vaginitis. In a recent study, 66.7% of vaginal swabs were diagnosed as normal flora, 10.4% as intermediate flora, and 22.9% as pathogenic flora. Aerobic vaginitis was the primary causative agent for vaginal infections, which E. coli and E. faecalis18 mostly dominated. S. aureus is also one of the common causative agents of vaginitis. A study in Iran has shown that the presence of Gram-positive bacteria was higher than Gram-negative bacteria. S. aureus appeared in 57.33% of the samples, followed by E. coli, 25.33 %19.
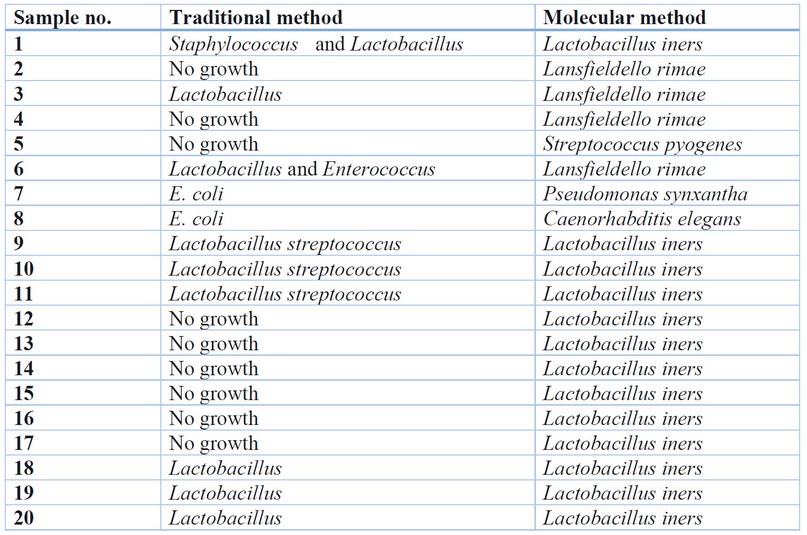
Table 1. Results obtained from traditional and molecular-based methods. Molecular-based methods were done in triplicates. When no pathogens were detected, the normal flora was listed.
Recombinant DNA method for bacterial identification
Amplification of the 16S rRNA gene from the genomic DNA isolated directly from vaginal swab samples showed that all samples amplified the 1495bp fragment of the 16S rRNA gene, as shown in Figure 1. This was expected as the vagina is not a sterile environment, and many normal flora are present; therefore, when using bacterial universal primers, all members of the vaginal microbiome will be amplified 20. In our recent publication, we showed that molecular identification by direct 16S rRNA improves the standards of identification in specimens that are known to be sterile, particularly blood and cerebrospinal fluid 9. However, direct amplification followed by sequencing for vaginal swabs is not appropriate in this case because this method will generate mixed amplicons, which must be separated before sequencing. One way of separating mixed sequences is via TA cloning21.
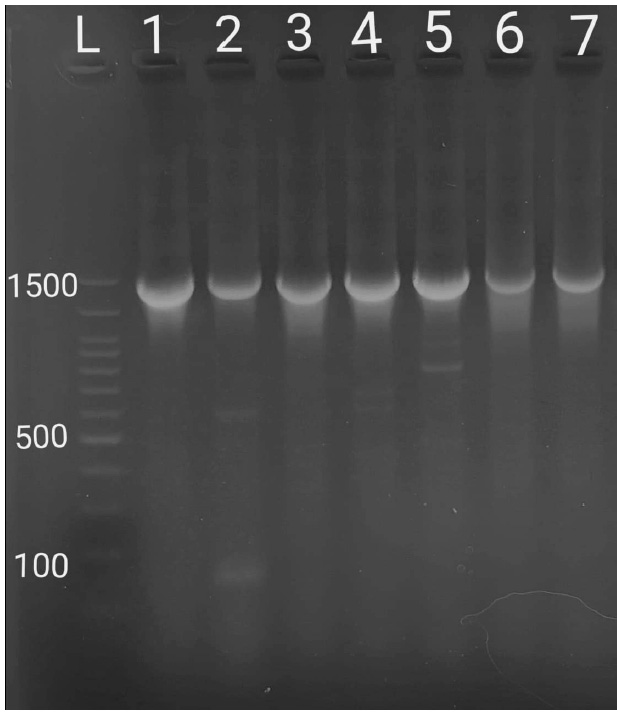
Figure 1. Amplification of 16S rRNA gene from a selection of bacterial genomic DNA isolated from vaginal swab samples. L: 100bp DNA ladder, 1-9: random samples selected.
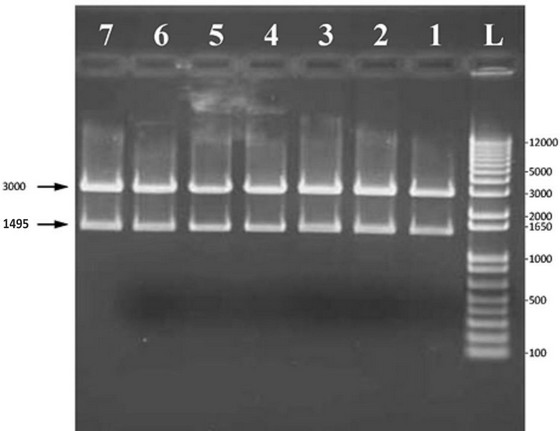
Figure 2. Digestion of pGEM-T easy vector containing 16S rRNA gene using EcoRI. L: 1Kb plus DNA ladder, 1-7: Plasmid DNA was digested with EcoRI from random samples that were selected and isolated from white colonies.
TA cloning results showed that the nine samples that did not show growth using traditional methods produced white colonies on LB agar plates containing ampicillin and X-gal. Digestion of plasmids from white colonies using EcoRI showed that they contain a 1495bp fragment (figure 2). Sequencing of plasmids isolated from white colonies showed that the vaginal swabs with no growth contained the normal flora Lactobacillus iners and the pathogens Streptococcus pyogenes, Pseudomonas synxantha and Lancefieldello rimae. These results come in agreement with a study by Mattei et al. as they found that the majority of the vaginal swabs contained the genus Lactobacillus, followed by Prevotella, Streptococcus, Ruminococcaceae and finally, Coriobacteraceae 22.
Lancefieldella is a strictly anaerobic bacterium that was formerly known as Atopobium. This lactic acid-producing bacteria was shown to be resistant to some antibiotics, particularly clindamycin, penicillin, moxifloxacin, and metronidazole 23. Atopobium vaginae is a member of this genus and were first isolated from a vaginal swab of a healthy person 24; however, to our knowledge, the species Lancefieldella image has not been previously isolated from vaginal swabs, this is probably because when using traditional methods cultures are usually incubated aerobically. In contrast, by using TA cloning method it is possible to detect all types of bacteria regardless of their type of nutrition or cultivation requirements. Even though Lancefieldella rimae is a lactic acid-producing bacteria, this bacterium has been shown to cause oral and dental infections, abdominal wound infections and tubo-ovarian abscesses 25. The detection of Lancefieldella rimae in vaginal swab samples could result from tubo-ovarian infections that may spread to the vaginal area. More studies are required to identify the role of this bacterium in pathogenesis.
Streptococcus pyogenes is a Gram-positive pathogen customarily known to infect mucosal membranes and skin, causing pharyngitis, pyoderma and impetigo. In immunocompromised patients, Streptococcal infections may be life-threatening. Therefore, antibiotic administration is mandatory 26. Due to its ability to colonize mucosal membranes, S. pyogenes is also a causative agent of vulvovaginitis. Special virulence regulators were expressed when present in the vagina 27. However, due to inefficient methods used for detection, the rates of streptococcal infections could be higher in adults. In a study to identify the antibiotic resistance spread among vaginitis in pregnant women, S. pyogenes was detected at the lowest ratio (11.6%) compared to S. aureus and E. coli, which were detected at (23.8% and 21.6%), respectively 28. A murine vaginal colonization model has been recently developed to detect group A and group B Streptococci to improve isolation and detection29. Our method using TA cloning successfully detected this pathogen and all other pathogens that might be found in the vagina. S. pyogenes was detected from a vaginal swab that did not show growth using traditional culture methods; this indicates the importance of molecular methods in identification.
Pseudomonas was also shown to be rare in vaginal swabs compared to other locations in the body. This was found in a study conducted in Nigeria, as it was found that Pseudomonas was mainly found in wound swabs (44%) followed by urine (30%) ear swabs (12%) and the least prevalence was seen in sputum and vaginal swab (0.67%)30. Our method detected Pseudomonas in one out of 20 tested samples (5%) and successfully identified the species as P. synxantha. Detection of species by our method is another advantage that can be added to the method understudy and would improve the results hospitals give if such methods were involved.
CONCLUSIONS
As technological methods improve, new methods for bacterial identification are introduced. This study attempts to introduce recombinant DNA technology in medical laboratories. Results showed higher accuracy in bacterial detection from specimens that are non-sterile of microbes, particularly vaginal swabs. Using TA cloning method for bacterial detection from vaginal swabs successfully detected both normal flora and pathogenic microbes isolated by traditional methods and other microbes not detected by traditional methods. Molecular tools should be considered by diagnostic laboratories worldwide for better health quality.
REFERENCES
1. Muhamad Rizal, N. S.; Neoh, H. M.; Ramli, R.; A/LK Periyasamy, P. R.; Hanafiah, A.; Abdul Samat, M. N.; Khor, B. Y. Advantages and limitations of 16S rRNA next-generation sequencing for pathogen identification in the diagnostic microbiology laboratory: perspectives from a middle-income country. Diagnostics, (2020). 10(10), 816.
2. Makdasi, E.; Atiya-Nasagi, Y.; Gur, D.; Zauberman, A.; Schuster, O.; Glinert, I.; Laskar, O. An improvement in diagnostic blood culture conditions allows for the rapid detection and isolation of the slow growing pathogen Yersinia pestis. Pathogens, (2022). 11(2), 255.
3. Abayasekara, L. M.; Perera, J.; Chandrasekharan, V.; Gnanam, V. S.; Udunuwara, N. A.; Liyanage, D. S.; Ilango, J. (2017). Detection of bacterial pathogens from clinical specimens using conventional microbial culture and 16S metagenomics: a comparative study. BMC infectious diseases, 17, 1-11.
4. Kalghatgi, A. T.; Praharaj, A. K.; Sahni, A. K.; Pradhan, D.; Kumaravelu, S.; Prasad, P. L.; Nagendra, A. (2008). Detection of bacterial pathogens in cerebrospinal fluid using restriction fragment length polymorphism. Medical Journal Armed Forces India, 64(1), 29-32.
5.Tunkel, A. R.; Scheld, W. M. Acute bacterial meningitis. The Lancet, (1995). 346(8991-8992), 1675-1680.
6. La Scolea Jr, L. J.; Dryja, D. Quantitation of bacteria in cerebrospinal fluid and blood of children with meningitis and its diagnostic significance. Journal of Clinical Microbiology, (1984). 19(2), 187-190.
7. Bonilla-Silva, E., & Dietrich, D. (2011). The sweet enchantment of color-blind racism in America. The ANNALS of the American Academy of Political and Social Science, 634(1), 190-206.
8. Grif, K., Heller, I., Prodinger, W. M., Lechleitner, K., Lass-Flörl, C., & Orth, D. (2012). Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples using a commercial 16S rRNA broad-range PCR and sequence analysis. Journal of clinical microbiology, 50(7), 2250-2254.
9. Khaleel, A. M.; Faisal, R. M.; Altaii, H. A. Molecular methods are more efficient than traditional methods in identifying bacteria from blood and cerebrospinal fluid samples. Malaysian Journal of Microbiology, (2023). 19, 2.
10. Gwinn, M.; MacCannell, D.; Armstrong, G. L. Next-generation sequencing of infectious pathogens. Jama, (2019). 321(9), 893-894.
11. Vandepitte, J., Verhaegen, J., Engbaek, K., Piot, P., Heuck, C. C., Rohner, P., & Heuck, C. C. (2003). Basic laboratory procedures in clinical bacteriology. World Health Organization.
12. de Melo Oliveira, M. G., Abels, S., Zbinden, R., Bloemberg, G. V., & Zbinden, A. (2013). Accurate identification of fastidious Gram-negative rods: integration of both conventional phenotypic methods and 16S rRNA gene analysis. BMC microbiology, 13, 1-10.
13. Abdulrazzaq R, Faisal R. Efficiency of Hichrome Enterococcus faecium Agar in the Isolation of Enterococcus spp. and other Associated Bacterial Genera from Water. Journal of Life and Bio Sciences Research, (2022). 3(01), 01-06.
14. Faisal, R. M. Understanding the role of Dibenzofuran 4, 4a dioxygenase reveals a silent pathway for biphenyl degradation in Sphingomonas wittichii RW1 and helps in engineering dioxin degrading strains (Doctoral dissertation, Rutgers University-School of Graduate Studies). (2019).
15. Sambrook, J. Molecular cloning: a laboratory manual. Synthetic oligonucleotides. (1989).
16. Giordani, B.; Abruzzo, A.; Parolin, C.; Foschi, C.; Laghi, L.; Marangoni, A.; Vitali, B. Prebiotic Activity of Vaginal Lactobacilli on Bifidobacteria: from Concept to Formulation. Microbiology Spectrum, (2023). e02009-22.
17. Pino, A.; Rapisarda, A. M. C.; Vaccalluzzo, A.; Sanfilippo, R. R.; Coman, M. M.; Grimaldi, R. L.; Verdenelli, M. C. Oral Intake of the Commercial Probiotic Blend Synbio® for the Management of Vaginal Dysbiosis. Journal of Clinical Medicine, (2023). 12(1), 27.
18. Salinas, A. M.; Osorio, V. G.; Pacha-Herrera, D.; Vivanco, J. S.; Trueba, A. F.; Machado, A. Vaginal microbiota evaluation and prevalence of key pathogens in ecuadorian women: an epidemiologic analysis. Scientific reports, (2020). 10(1), 1-18.
19. Ghiasi, M.; Fazaeli, H.; Kalhor, N.; Sheykh-Hasan, M.; Tabatabaei-Qomi, R. Assessing the prevalence of bacterial vaginosis among infertile women of Qom city. Iranian journal of microbiology, (2014). 6(6), 404.
20. Diop, K.; Dufour, J. C.; Levasseur, A.; Fenollar, F. Exhaustive repertoire of human vaginal microbiota. Human microbiome journal, (2019). 11, 100051.
21. Hooper, S. J.; Crean, S. J.; Fardy, M. J.; Lewis, M. A.; Spratt, D. A.; Wade, W. G.; Wilson, M. J. A molecular analysis of the bacteria present within oral squamous cell carcinoma. Journal of medical microbiology, (2007). 56(12), 1651-1659.
22. Mattei, V.; Murugesan, S.; Al Hashmi, M.; Mathew, R.; James, N.; Singh, P.; Tomei, S. Evaluation of methods for the extraction of microbial DNA from vaginal swabs used for microbiome studies. Frontiers in Cellular and Infection Microbiology, (2019). 9, 197.
23. Wolf, L. J.;Stingu, C. S. Antimicrobial Susceptibility Profile of Rare Anaerobic Bacteria. Antibiotics, (2023). 12(1), 63.
24. Jovita, M. R.; Collins, M. D.; Sjödén, B.; Falsen, E. Characterization of a novel Atopobium isolate from the human vagina: description of Atopobium vaginae sp. nov. International Journal of Systematic and Evolutionary Microbiology, (1999). 49(4), 1573-1576.
25. Lamont, R. F.; Sobel, J. D.; Akins, R. A.; Hassan, S. S.; Chaiworapongsa, T.; Kusanovic, J. P.; Romero, R. The vaginal microbiome: new information about genital tract flora using molecular based techniques. BJOG: An International Journal of Obstetrics & Gynaecology, (2011). 118(5), 533-549.
26. Fiedler, T.; Köller, T.; Kreikemeyer, B. Streptococcus pyogenes biofilms—formation, biology, and clinical relevance. Frontiers in cellular and infection microbiology, (2015). 5, 15.
27. Cook, L. C.; Chatterjee, N., Li, Y.; Andrade, J.; Federle, M. J.; Eichenbaum, Z. Transcriptomic analysis of Streptococcus pyogenes colonizing the vaginal mucosa identifies hupY, an MtsR-regulated adhesin involved in heme utilization. Mbio, (2019). 10(3), e00848-19.
28. Mohammed, A. B.; Hamadamin, H. A. Antibiotic sensitivity of high vaginal swabs from asymptomatic pregnant women. Diyala Journal of Medicine, (2021). 20(1), 70-75.
29. Verstraelen, H.; Verhelst, R.; Vaneechoutte, M.; Temmerman, M. Group A streptococcal vaginitis: an unrecognized cause of vaginal symptoms in adult women. Archives of gynecology and obstetrics, (2011). 284, 95-98.
30. Adejobi, A.; Ojo, O.; Alaka, O.; Odetoyin, B.; Onipede, A. Antibiotic resistance pattern of Pseudomonas spp. from patients in a tertiary hospital in South-West Nigeria. Germs, (2021). 11(2), 238.
Received: 25 June 2023/ Accepted: 26 August 2023 / Published:15 September 2023
Citation: Khaleel, A. M.; Faisal, R. M.; Altaii, H. A. (2023). Using recombinant DNA technology in bacterial identification from vaginal swabs. Revis Bionatura 2023;8 (3) 113 http://dx.doi.org/10.21931/RB/2023.08.03.113
