2023.08.03.94
Files > Volume 8 > Vol 8 No 3 2023
Bioremediation by bacteria isolated from water contaminated with hydrocarbons
1,2,3 Department of Microbiology, College of Science, Al-Mustansiriyah University, Iraq.
*Corresponding authors: Email: [email protected],
, Mobile: +964 7718051974
Available from: http://dx.doi.org/10.21931/RB/2023.08.03.94
ABSTRACT
Oil pollution is currently a global problem. However, an oil-contaminated ecology is rich in microorganisms that may utilize petroleum oil and hydrocarbons for growth, feeding, and metabolic processes. In the present study, fifty polluted water samples were collected from five stations (ten samples each) in the Al-Fahama oil refinery in eastern Baghdad. The water contamination parameters of these collected water samples were detected. Then, the percentage of water contamination with some heavy metals (zinc, lead, and cadmium) and radioactive elements (uranium, cesium and actinium) was measured. The proportions of these elements were compared within their limits permitted by the World Health Organization (WHO). Fifty-nine bacterial isolates were isolated from polluted water, and 24 isolates of them succeeded in analyzing crude oil. The results of the current study showed that seven isolates belong to the genus Citrobacter amalonaticus (29.16%), six isolates belong to Enterobacter cloacae (25%), three isolates belonged to both Pseudomonas aeruginosa (12.5%) and Ochrobacterum anthropi (12.5%), and human Ochrobacterum. With a percentage of 12.5%, two isolates of Serratia marcescens (8.3%) and one isolate of each Pseudomonas fluorescens, Serratia fonticola, and Burkholderia pseudomallei (4.16%) of each. The optimum of some conditions for the decomposition process was determined in terms of (pH, temperature and crude oil concentration) and the results showed that the optimum degradation conditions were 35°C at pH equal to 7.5 in the presence of 2% of crude oil. Several experiments were conducted to determine the most efficient isolates for oil analysis. Burkholderia pseudomallei and Pseudomonas fluorescens are the most active bacterial species in their oil degradation. Genes responsible for hydrocarbon analysis were revealed in twenty-four bacterial isolates using a polymerase chain reaction (PCR) assay. The results showed that the ALKB gene (alkane hydroxylase) was observed in all bacterial isolates that succeeded in analyzing crude oil with a percentage equal to 100%, NahAc gene (naphthalene dioxygenase) has been recorded in four isolates (16.7%), these four bacterial isolates were Burkholderia pseudomallei, Pseudomonas aeruginosa, Ochrobacterum anthropic, and Pseudomonas fluorescens. Generally, the isolation rate of both C. amalonaticus and E. cloacae isolates was higher than in other studies, which may be due to the hydrocarbon pollution in isolation; both B. pseudomallei and P. fluorescens isolates were the highest active bacterial species in their oil degradation. Genetic results showed that the AlkB gene was the domain compared with other degradation genes used in the current study, followed by NahAc gene.
Keywords: Bioremediation, heavy metal, B. pseudomallei, hydrocarbons, crude oil
Because of their energizing and carbon needs for development and reproduction, in addition to the need to relieve stress response caused by the presence of hydrocarbons in the microbial mass environment, most hydrocarbons encountered in the surroundings are eventually degraded or hydrolyzed by indigenous bacteria 1, Non-regulated oil sludge removal, oil extraction, refining, and leaks during storage and transport all resulted in considerable soil pollution and aliphatic and aromatic hydrocarbon buildup in the environment, Since many petroleum hydrocarbons may be detrimental to living creatures, the elimination of these organic contaminants has received a lot of attention 2, Hydrocarbon pollution is a big concern because of its wide-ranging effects on all forms of life. Pollution produced by increasing crude oil consumption is ubiquitous due to its extensive use and the attendant transportation and disposal concerns. 3; the composition of crude oil is a complex mixture of aliphatic, aromatic, and aromatic particles. Heavy elements are naturally present in the soil, and their level rises due to human activities, including oil refining and insecticide use. Several places have such high quantities of metalloids and heavy metals that they have seriously affected the natural ecosystem. Heavy elements and metalloids reduced microbe activity, making them unsuitable for petroleum breakdown and hence less efficient 4, More than 98% of natural raw rubber comprises hydrocarbon polymers, a chain-like structure of many atoms connected. The types of chemical bonds that bind the particles of the constituent molecules affect the chemical system of hydrocarbons 5. Hydrocarbon contamination happens due to toxic organic compounds, petroleum, and pesticides, posing a severe environmental threat. Contamination produced by petroleum hydrocarbons is a source of concern because these are toxic to a variety of biological types6. Crude oil pollution is ubiquitous due to its widespread use, the associated disposal procedure, and unintentional spills. Petroleum is a complex combination of various hydrocarbons with high and low molecular weight. Petroleum combines alkanes, alkenes, and homo- and heterocyclic naphthenes, saturated and branched 7. Heavy metal and other compounds like N, S, and O are aromatics made up of heteroatoms. Ethers, carboxylic acids, and significant aromatic compounds are all examples of resins, asphaltenes, and naphthene-aromatics are examples of hydrocarbons with diverse active groups 8, 10. This study aimed to identify the bacterial isolates in the pollutant water of hydrocarbons and to determine their ability to degrade the hydrocarbons as well as the optimum conditions of the degradation process
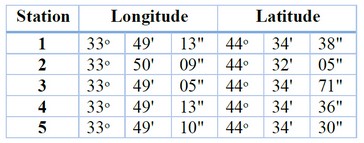
Sample collection: Fifty water samples of polluted water were taken from five stations located in Al-Fahama east of Baghdad (Al-Quds station as a model); these samples were collected in sterilized one-litter containers (from 30 ‐ 50 cm depth), Global positioning system parameters (GPS) of the isolation areas were illustrated in table (1).Table 1. Global positioning system parameters (GPS) of the studied area

Figure 1. Satellite image of the study area
Determination of the element levels in water samples
A: Determine some heavy elements levels in water samples: The levels of three selected heavy metals (cadmium, lead, and zinc) were examined and carried out in general company for foodstuff trade/food quality control division in Baghdad by using flame atomic absorption spectrometer in laboratories of the College of Science/ Al-Mustansiriyah University 10.
B: Determine some radioactive element levels in water samples: Levels of uranium, cesium, and actinium were determined in collected water samples as radioactive elements to determine if there is any presence in the area. A high-purity germanium radiation detector (HPGe) was used to determine the radioactive levels in the general company for foodstuff trade/food quality control division in Baghdad in laboratories of examination and audit division of the Ministry of Commerce 10.
Culture media: BH: Bushnell- Haas medium was prepared as illustrated in the previous study 11.
Bacterial identification: identification of bacteria according to their culture, morphological, and biochemical test, which include (Gram stain, Catalase, Oxidase, Indole, Methyl red, Citrate, Urease, H2S, motility, and VP test, as well as VITEK2 system 10.
Determine the biodegradation ability of isolated bacteria.
A. detection by using crude oil agar plates: A 0.1ml of crude oil was diffused by the glass spreader on BH media, which was prepared, then the bacterial isolates were cultured separately on a media, and the plates were incubated at 37 ºC for five days.
B. Measure the dissolved oxygen values: Dissolved oxygen values (DO) of 20 ml of the BH broth media containing the isolated bacterial strains were measured by using DO meter daily for 15 days to determine the (COD) values in the culture 12.
C. DCPIP photosensitizer method: Bacterial isolates were inoculated into 100 mL of nutrient broth and cultured at 37°C in a shaker incubator with 150 rpm for 24 hours. The cultures were used as a source of hydrocarbon degradation inoculum. One hundred ml of BH media were made with one percent diesel as the sole carbon source. DCPIP (2,6-Dichlorophenolindophenol) was added at a concentration of 20 mg/l in the media. The media was sterilized for 15 minutes at 121°C, and each set of experiments received 1 ml, which was incubated at 150 rpm for 14 days; tubes without bacterial suspension and DCPIP solution were used and considered as negative and positive controls. Changing the color of the tubes from blue to colorless was a visual detection of the ability of bacterial isolates to degrade the hydrocarbons, and this color change can be measured spectrophotometrically at 630 nm by ELISA reader spectrophotometer 12.
D. Gravimetric analysis assay
Gravimetric analysis assay can be used to estimate the amount of hydrocarbons degrading biologically by bacterial isolates. 50 ml of BH broth culture of each strain was transferred separately in a 250 ml conical flask 1N HCl was added to the culture to stop the bacterial activity. Then 50 ml of petroleum ether and acetone (1:1) was added. After that, the mixture was transferred to the separating funnel and mixed well for 15 minutes. After another 15 minutes of sitting, the solution was divided into three phases: an organic phase on top, an aqueous phase in the middle, and a cell biomass phase on the bottom. The upper solvent, which contains the oil, was separated from the two other phases, and then it was transferred to a beaker; after measuring the weight of the empty beaker, and kept in the oven at 45-50º C till the solvent was evaporated, the final weight of the beaker was measured, and thus The ratio of degraded crude oil was calculated by using the formula below.
Crude oil degraded % = (weight of petrol degraded / weight of petrol present)X 100
Where the weight of petrol degraded = original weight of petrol - weight of residual petrol obtained after evaporating the solvent 12.
Determining the optimum parameters for the bacterial decomposition of hydrocarbons
A: Temperature degree: One hundred ml of BH broth was transferred into a 250 ml flask and then sterilized by autoclaving (121 º C for 15 min). After that, 1% of the crude oil was added to the flask with one ml of the isolated bacterial suspension (0.5 McFarland solution), and the flask was incubated in a shaker incubator at different temperatures (25, 30, 35, and 40 ºC). The results were recorded daily for five days to determine the optimum temperature for the hydrocarbon biodegradation 13.
B. PH value: One hundred ml of BH broth (it was illustrated in 1.3.1.) was transferred into a 250 ml flask with different pH values (6, 7, and 8), and then it was sterilized by autoclaving (121 º for 15 min). After that, 1% of the crude oil was added to the flask with 1 ml of bacterial suspension (0.5 McFarland solution), and the flask was incubated in a shaker incubator at 37 ºC. The results were recorded daily for five days to determine the optimum temperature for the hydrocarbon biodegradation 14.
C. Crude oil concentration: One hundred ml of BH broth was transferred into a 250 ml flask and then sterilized by autoclaving (121 º for 15 min). After that, different concentrations of crude oil (1%, 2%, and 3%) were added to the flask with one ml of bacterial suspension (0.5 McFarland solution), and the flask was incubated in a shaker incubator at 37 ºC. The results were recorded daily for five days to determine the optimum crude oil concentration 14.
Detection of degradation genes in bacterial isolates: PCR was used to detect three catabolic genes that encode enzymes involved in degradation pathways in purified DNA. alkB, C320, PAH, C120 and Nah genes were amplified using the primers illustrated in Table 2. The resulting products were tested on an agarose gel and stained with ethidium bromide to confirm gene amplification. PCR was programmed of three genes (AlkB, Nah, and PAH) into four stages: primary denaturation stage of one cycle of pre-denaturation at 95º for 5 minutes. This amplification stage of thirty-five cycles consists of 94º for 40 sec, 55º for 40 sec, 72º for 1 minute, followed by the final extension stage of one cycle as 72º for 10 minutes, while the PCR program of both C230 and C120 genes was designed as 5 min as initial denaturation at 95°C, thirty-five cycles at 94° for 20 sec, 63° for 30 sec, and 45 s at 72°C, and final elongation for 10 min at 72°C. PCR products were then transferred and resolved separately by electrophoresis on agarose gel. The presence of a clear band in the product of any test using ultraviolet light referred to the presence of the required gene in the bacterial isolate 13.

Table 2. List of primers which are used in this current study.
RESULTS
Results of water samples analysis
A: Results of heavy metals levels in water samples: The results of heavy metals (cadmium, lead, and zinc) estimations in water samples showed that zinc was recorded as a higher percentage compared with lead and cadmium, the zinc average was recorded as 1.168 mg l-1 mg l-1, and the highest value was 1.41 mg l-1, and the lowest value was 0.695 mg l-1. The results showed that the lead element came in second place, with an average of 0.467 mg l-1, where its concentrations ranged between 0.06 to 0.36 mg l-1. Finally, cadmium concentrations were recorded as the lowest of the selected heavy metals, ranging from 0.002 to 0.0003 mg l-1, with 0.001 as an average, as illustrated in Table (3).
B: Results of radioactive elements levels in the water sample
The energy of radioactive elements in water samples was zero in all the collected water samples, which indicates the absence of recent or previous radioactive contamination in the study area.
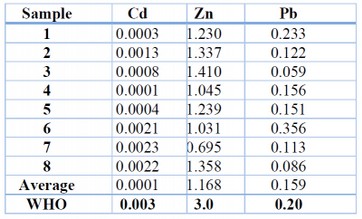
Table 3. Results of heavy metals levels in water samples
Results of bacterial isolation and identification:
Fifty-nine bacterial samples were isolated from 60 water samples collected from oil refining wells in the Al-Fahama area; the percentage of the bacterial isolates from water samples was higher than the other previous studies; it was equal to 98.3 %, which was recorded as high value may due to the bacterial contamination in the study area, as well as the presence of many bacterial isolates that can withstand or degraded the hydrocarbons 15. Out of the total isolates, twenty-four bacterial isolates succeeded in analyzing oil and using it as an energy source, where the percentage of isolation of oil-consuming bacteria was 40%. Bacterial isolates were identified by observing phenotypic characteristics on different used media, investigating microscopic characteristics with Gram stain, and performing biochemical and VITEK 2 tests. The results showed that the isolated bacteria represented several bacterial genomes. The current study results recorded that seven isolates (29.16 %) belonged to the genus Citrobacter amaloonaticus, six isolates (25 %) belonged to Enterobacter cloacae bacteria as a second common collected isolates, three isolates of both Pseudomonas aeruginosa and Ochrobacterum anthropic bacteria, with percentage equal to 12.5 %. Moreover, the isolation results showed the presence of four other bacterial genera as one isolate of each, with a percentage equal to 4.16% for each; these bacterial isolates were (Pseudomonas fluorescens, Serratia marcescens, Serratia fonticola and Burkholderia pseudomallei), as shown figure (2).
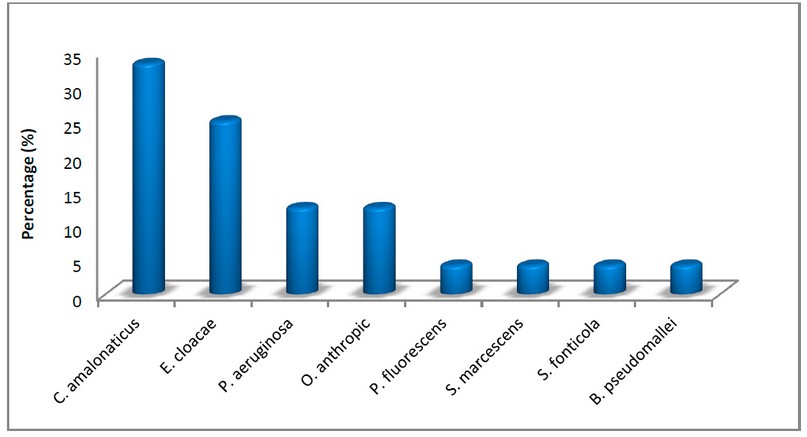
Figure 2. Isolation percentages of crude oil.
Results of the optimum conditions of bacterial decomposition of hydrocarbon
A. Determine the optimum temperature: When biodegradation of crude oil is carried out on BH broth and solid media and incubated at four different temperatures (25, 30, 35 and 40) ºC, the results showed that the optimum temperature for the decomposition of crude oil is 35 ºC. The results we obtained agreed with the number of studies that showed that the biomass of oil-degrading Pseudomonas aeruginosa bacteria reaches a maximum of 35 ºC. The biomass decreases at 25, 30 and 40 ºC 16 because the temperature affected hydrocarbon metabolism and the hydrocarbon's physical and chemical nature considered 35 ºC the best temperature for analyzing hydrocarbon compounds 17.
B. Determine the optimum pH value: When cultured bacteria in a BH broth media with crude oil, the pH was adjusted to find the best grade for crude oil breakdown; the optimal pH level was shown to be 7.5 according to the results of the current study. The pH of a bacterial cell influences its metabolism, which in turn influences hydrocarbon degradation, as well as biomass production 18.
C. Determine the optimum crude oil concentration: To find out the appropriate concentration to increase the biodegradation process, this contains different concentrations of crude oil. After conducting several experiments, it was found that the best concentration of biodegradation of crude oil was 2%; when the experiment was performed at a concentration greater than 2%, we discovered that biodegradation was less efficient.
Quantitative results of the bacterial biodegradation to crude oil: The biodegradation rate of crude oil by the isolated and identified bacterial isolates was performed by using four different methods:
A. Result of measure of dissolved oxygen demanded method: It was noted that the Burkholderia pseudomallei bacteria achieved the most considerable significant difference from the Control, which reached 2.04, as it recorded the highest oxygen consumption. Citrobacter alamoaticus bacteria recorded the least significant difference from Control, which amounted to 4.13, where the percentage of moral error was 0.583, as these bacteria had the lowest oxygen consumption, see Figure 3.
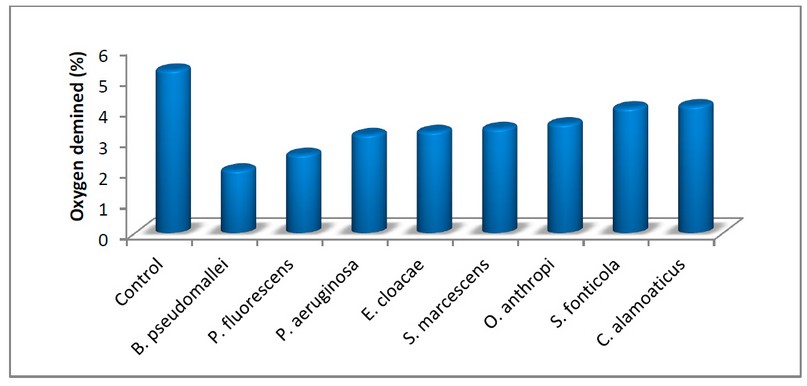
Figure 3. Results of bacterial dissolved oxygen consumption.
Results of crude oil biodegradation by using agar plates
The results showed that the most efficient and fastest degrading isolates belong to the Burkholderia pseudomallei bacteria due to the presence of high significant differences between them and the Control, which amounted to 0.34, where the ratio of the standard line was 0.24, where the highest moral difference was achieved. Serratia fonticola bacteria showed the least significant difference, which amounted to 0.81, and the standard error ratio was 0.38.
C. Result of DCPIP electron acceptor method: Using the color reagent, B. pseudomallei, E. Cloacae and P. aeruginosa showed a noticeable color change after 120 hours of incubation. C. alamoaticus, O. anthropic and P. fluorescens noticeable discoloration appears after 168-192 hours. S. fonticola, S. marcescens, there is no noticeable discoloration observed in the dye, as shown in Figure (4). The study used the color change of the dye Dichlorphenol Indophenol to determine the efficiency of bacteria in analyzing hydrocarbon compounds, Returns the dye's color disappearing as a result of absorbing an electron arising from the oxidation of hydrocarbons by bacteria 19.
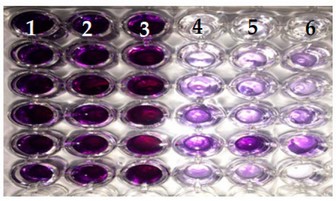
Figure 4. Result of DCPIP electron acceptor method. Column one (blue color) represents the Control, the second two columns recorded as negative results, while columns four to six (colorless) represent positive results.
D. Result of gravimetric analysis assay: All the isolates could degrade the crude oil, and the degradation ranged from 1.14 to 2.43 gm. Moreover, the results showed that P. aeruginosa recorded the highest degradation compared with other tested bacteria, with 2.43 gm. The results also showed that the highest percent of degradation was recorded on the third day of incubation, as shown in Figure (5).
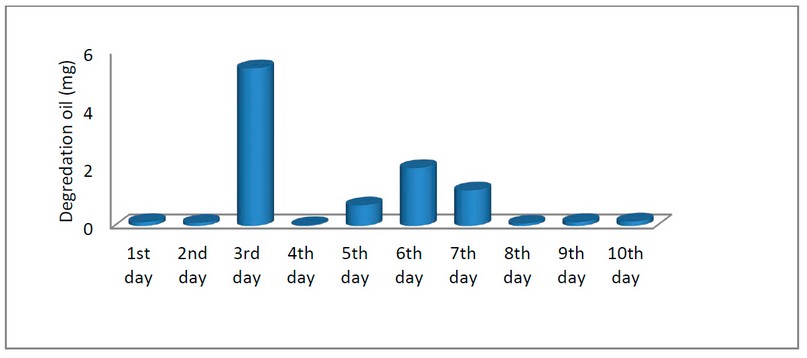
Figure 5. Results of crude oil biodegradation weight of the isolated bacteria.
Detection of hydrocarbon degradation genes in the isolated bacteria: Detection of the presence of genes responsible for hydrocarbon analysis was revealed in twenty-four bacterial isolates that succeeded in analyzing crude oil by using polymerase chain reaction (PCR) assay. The results showed that ALKB gene (alkane hydroxylase) was observed in all bacterial isolates that succeeded in analyzing crude oil with a percentage equal to 100%. NahAc gene (naphthalene dioxygenase) has been recorded in four isolates with a percentage equal to 16.7%; these four bacterial isolates were Burkholderia pseudomallei, Pseudomonas aeruginosa, Ochrobacterum anthropic, and Enterobacter cloacae), see figure 6. On the other hand, TodcI, C230, and Cat23 genes do not appear in all isolated bacteria. Table (4) demonstrates the appearance of the studding genes in isolated bacteria.
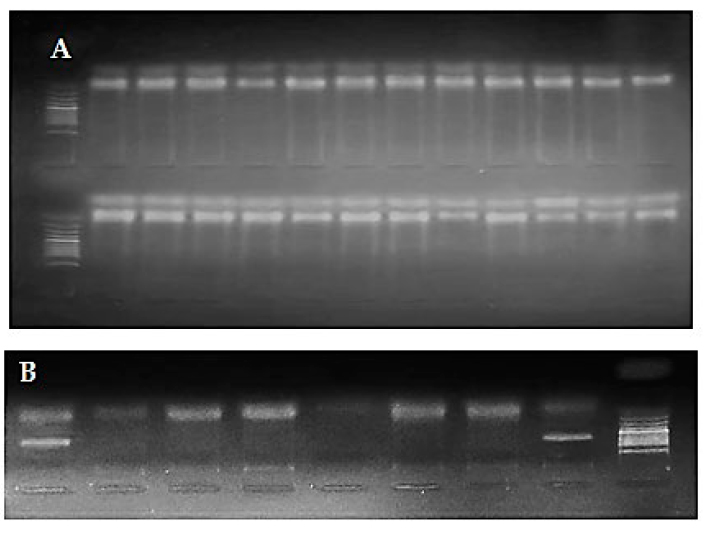
Figure 6. Occurrence of bacterial biodegradation results. Where A: ALKB gene, B: NahAC genes
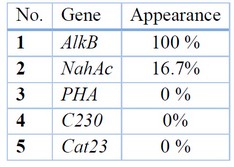
Table 4. Appearance of degradation genes in bacterial isolates.
The results showed that water samples collected in the current study were within the permissible limit of heavy metals that were chosen compared with acceptance levels of WHO 20. Although many local studies illustrated the high level of cadmium in the Najaf Sea in Iraq due to the association of its presence with the wastes presented for oil refining or places of extraction of fossil fuels 21, another local study found that the concentration of cadmium and lead higher in Kirkuk irrigation project 22. The results of the isolation of bacteria matched the isolation carried out in many previous studies 23, while the bacterial isolation percentage contrasted with the percentage reached by scientist 24, which was 23%. The difference in isolation rates is due to the difference in the size of the sample or the different conditions of isolation. Studies have shown that Gram-negative and specifically Citrobacter amalonaticus, ability to analyze hydrocarbon compounds is due to the presence of an efficient enzymatic system capable of breaking down these compounds and using them as an energy source 25, its high analytical ability of hydrocarbons may be due to its possession of specific genes or plasmids, and it may contain in its cell walls 26. Bacteria can break down hydrocarbons because they have an enzyme that consists of the enzymes oxidase and peroxidase, which bind oxygen with these compounds, which helps degrade them 27.
According to a number of studies, the optimum temperature for oil degradation is 35-40 ° C, lower temperatures impact the physical state of crude oil by reducing alkane volatilization, which leads to a rise in the viscosity of the oil and thus makes the biological analysis process more difficult 28, for different numbers of bacterial groups. At 20-30 ºC, pseudomonas aeruginosa bacteria destroyed 85 % of the crude oil 29.
Oxygen demanded is the amount of oxygen a microorganism uses to carry out the biodegradation process 30. The low percentage of oxygen is due to the ability of bacteria to consume it in the biodegradation process of hydrocarbons, as aerobic bacteria depend on the hydrogenase enzyme in the oxidation of hydrocarbons, and this explains the inverse relationship between the biodegradation process and the percentage of dissolved oxygen 31. Many previous studies have shown that Burkholderia sp is one of the most suitable isolates for its ability to analyze toluene, naphthalene, kerosene, and alanine. The intermediate results of the analysis process were non-toxic. Burkholderia has oxygenase enzymes for the oxidation of substituted carbon 32. Studies have observed the ability of both Pseudomonas aeruginosa and Burkholderia pseudomallei bacteria to decompose hydrocarbon compounds due to their possession of the enzyme 2,3-dioxygenase, which is responsible for inserting oxygen between the carbon chains, which leads to the splitting of benzene and the formation of intermediate compounds 33, Studies have shown that the Burkholderia pseudomallei bacteria contain a binary enzyme system capable of decomposing hydrocarbons with high efficiency (monooxygenases and dioxygenases), It includes a specific enzyme, Dihydroxycyclohexadienecarboxylate dehydrogenase, carbon, which makes it more efficient in decomposing benzene and tolenene and using it as a carbon source, A study was conducted to choose the best isolates for the hydrolyzed crude oil, as it showed that P. aeruginosa and B. cereus bacteria are among the best isolates of the oil that are attracted by the analyzes for their secretion into protein materials capable of cracking the chains between the carbon atoms 29. A study was conducted to analyze the rDNA of Burkholderia pseudomallei bacteria, and it was found that the ability of this bacteria to analyze complex compounds in the environment as a source of carbon is due to its possession of 2,2 dehalogenase (2,2-DCP) 34.
The gravimetric analysis assay method did not give tangible and accurate results to know the amount of residual hydrocarbons, and significant differences were observed between bacteria types during 10 days. Because of its properties, crude oil is one of the significant environmental pollutants capable of inflicting substantial harm to humans and the ecosystem. Prolonged exposure and high oil concentrations may result in liver or renal dysfunction, bone marrow damage, and an increased cancer risk 35.
Recent research suggests that bacteria that degrade hydrophobic chemicals like PAHs and alkanes have unique physiological characteristics. Sphingomonads have been shown to have a wide range of PAH metabolic capacities and to be engaged in the degradation of various refractory organic pollutants, including biphenyl and benzoate, pyrene. Furthermore, the breakdown of various PAH molecules by isolated bacteria appears through separate mechanisms 36. The catabolic genes can be used to track a bacterial strain's specific activities, The AlkB (alkane hydroxylase) gene encodes the enzyme monohydrogenase, The NahAc (naphthalene dioxygenase) gene encodes the enzyme dioxygenase, the enzymes monohydrogenase and dehydrogenase are responsible for the catalyze of aliphatic and PAHs compounds 32. The majority of bacteria that degrade PAHs are Gram-negative, according to the study. The results of conventional features led to the identification of ASU-06 as Sphingomonas sp. The mono- and dioxygenases generated by bacteria were primarily responsible for the degradation of aliphatic and PAHs. As a result, six essential enzyme coding genes in S. koreensis strain ASU-06 were studied, including monooxygenase and dioxygenase. Monooxygenase (alkB and alkB1), dioxygenase (NahAc), and Catechol dioxygenase (C12O and C23O) genes have been discovered. One possible explanation is that the genes found are highly conserved among Gram-negative bacteria. However, no signal was seen because the PAH-RHD gene is conserved among Gram-positive bacteria. The naphthalene dioxygenase (NahAc) gene is fascinating as a PAHs degradation indicator since the enzyme produced by this gene destroys naphthalene and promotes the degradation of other PAHs compounds 36. Proteins or genes of 1-hydroxy-2-naphthoic acid dioxygenase have been found in Gram-negative and Gram-positive bacteria belonging to the genera Pseudomonades, Nocardioides, and Mycobacterium, The reports showed the positive detection of the presence of the gene NahAc indicating that bacteria are on the analysis of polycyclic hydrocarbons, the study showed the presence of the Nah gene in many genera of negative bacteria, such as Pseudomonas and Bolkhaderia. Because genes conserved in Gram-negative bacteria are present in Gram-positive bacteria, the distribution of NahAc gene among PAH-degrading bacteria implies the influence of horizontal gene transfer (HGT) 37. Finally, the molecular technique recommended the identification of different area, including biology fields
The results showed a high isolation rate of C. amalonaticus isolates compared with other results in the same area and the same thing for E. cloacae isolates. Also, the presence of crude oil should not be more than 2% as an optimum condition of biodegradation with pH close to natural and temperature more than room temperature, which differs from many other results that mentioned room temperature as an optimum temperature of crude oil degradation by environmental isolates. B. pseudomallei and P. fluorescens isolates were the most active bacterial species in their oil degradation. Genetic results showed that the AlkB gene was the domain compared with other degradation genes used in the current study, followed by the NahAc gene.
Funding: Self-Funding.
Acknowledgments:
At the end of this research, we must thank the University of AlMustansiriyah for all the equipment and facilities we used to do and finish that research. Also, we thank all authors who contributed to the design of the study, those who contributed to the acquisition, analysis, or interpretation of the data, and others who contributed to the writing of the manuscript.
Conflicts of Interest: we, as authors, have to declare that there is no conflict of interest.
1. Chow, J., Lee, S. M., Shen, Y., Khosravi, A., & Mazmanian, S. K. Host–bacterial symbiosis in health and disease. Advances in immunology. 2010; 107, 243-274.
2. Emoyan, O. O., Agbaire, P. O., Ohwo, E., & Tesi, G. O. Priority mono-aromatics measured in anthropogenic impacted soils from Delta, Nigeria: concentrations, origin, and human health risk. Environmental Forensics.2021;1-13.
3. Gao, H., Wu, M., Liu, H., Xu, Y., & Liu, Z. Effect of petroleum hydrocarbon pollution levels on the soil microecosystem and ecological function. Environmental Pollution. 2021;293, 118-124.
4. Ossai, I. C., Ahmed, A., Hassan, A., & Hamid, F. S. Remediation of soil and water contaminated with petroleum hydrocarbon: A review. Environmental Technology & Innovation. 2020;17, 100526.
5. Srivastava, M., Srivastava, A., Yadav, A., & Rawat, V. Source and control of hydrocarbon pollution. In Hydrocarbon Pollution and its Effect on the Environment. Intech Open. 2019;12, 243-274.
6. Guerra, A. B., Oliveira, J. S., Silva-Portela, R. C., Araujo, W., Carlos, A. C., Vasconcelos, A. T. R., et al. Metagenome enrichment approach used to select oil-degrading bacteria consortia for drill cutting residue bioremediation. Environ. Pollut. 2019;235, 869–880.
7. Varjani, S. J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017;223, 277–286.
8. Priyadarshanee, M., Mahto, U., & Das, S. Mechanism of toxicity and adverse health effects of environmental pollutants. In Microbial Biodegradation and Bioremediation. 2022; 33-53.
9. Abed, I.A., Hameed, A.T., Abdulilah, H.A.Q. The role of Glomus mosseae in the biochemical recommendation of contaminated hydrocarbon (black oil) soils Plant Archives. 2019;19, 322–326.
10. Mohammed F. AboKsour, Shatha A. Shafiq and Abbas H. Salman. Response of some pathogenic microorganisms to crude alkaloid of endophytic fungi isolated from Catharanthus roseus. Journal of Genetic and Environmental Resources Conservation. 2021;9(1):190-196.
11. Powell SM, Ferguson SH, Bowman JP, Snape I. Using real-time PCR to assess changes in the hydrocarbon-degrading microbial community in antarctic soil during bioremediation. Microbial Ecology. 2021;52(3):523–532.
12. Abd El-Latif Hesham, Asmaa M. M. Mawad, Yasser M. Mostafa, and Ahmed Shoreit. Biodegradation Ability and Catabolic Genes of Petroleum-Degrading Sphingomonas koreensis Strain ASU-06 Isolated from Egyptian Oily Soil. Biomed Res Int. 2021;73(2):148–159.
13. Hassanshahian, M., Zeynalipour, M. and Musa, F. Isolation and characterization of crude oil degrading bacteria from the Persian Gulf (Khorramshahr Provenance). Marine Pollution Bulletin. 2014;82, 39-44.
14. Varjani, S. J. Microbial degradation of petroleum hydrocarbons. Bioresour. Technol. 2017;23, 277–286
15. . Liu, L., Li, W., Song, W., & Guo, M. Remediation techniques for heavy metal-contaminated soils: Principles and applicability. Science of the Total Environment. 2018;633, 206-219.
16. Ellis, M., Altshuler, I., Schreiber, L., Chen, Y. J., Okshevsky, M., Lee, K., & Whyte, L. G. Hydrocarbon biodegradation potential of microbial communities from high Arctic beaches in Canada's Northwest Passage. Marine Pollution Bulletin. 2022; 174, 113288.
17. Mondal, M. H., Malik, S., Roy, A., Saha, R., & Saha, B. Modernization of surfactant chemistry in the age of gemini and bio-surfactants: a review. RSC advances. 2015;5 (112), 92707-92718.
18. Al-Hawash, A. B., Dragh, M. A., Li, S., Alhujaily, A., Abbood, H. A., Zhang, X., & Ma, F. Principles of microbial degradation of petroleum hydrocarbons in the environment. The Egyptian Journal of Aquatic Research. 2018;44 (2), 71-76.
19. Okafor, C. P., Udemang, N. L., Chikere, C. B., Akaranta, O., & Ntushelo, K. Indigenous microbial strains as bioresource for remediation of chronically polluted Niger Delta soils. Scientific African. 2021;11, 2793–2808
20. Mohammed F. AboKsour. Presence of Extended-Spectrum β-Lactamases Genes in E. coli Isolated from Farm Workers in the South of London. International Journal of Pharmaceutical Quality Assurance. 2018;9(1); 64-67.
21. Mahmood, I. Towards an effective enforcement of environmental criminal law: re-thinking sanctions for air pollution criminals in Iraq. University of Salford (United Kingdom). 2018.
22. Namuq, M. A. Studying of some heavy metals levels in water samples from Kirkuk Irrigation project of Tuz Khurmatu District, Iraq. Tikrit Journal of Pure Science. 2021;26(2), 48-52.
23. Vega, L., Jaimes, J., Morales, D., Martínez, D., Cruz-Saavedra, L., Muñoz, M., & Ramírez, J. D. Microbial Communities’ Characterization in Urban Recreational Surface Waters Using Next Generation Sequencing. Microbial Ecology. 2021;81(4), 847-863.
24. Mohamed, R. M. A. Investigation of Chemical Constituents of Ocimum basilicum Oil and its Antimicrobial Activity (Doctoral dissertation, Sudan University of Science and Technology). 2018.
25. Jacob, J. H., & Irshaid, F. I. Toluene biodegradation by novel bacteria isolated from polluted soil surrounding car body repair and spray painting workshops. Journal of Environmental Protection. 2015;6(12), 1417.
26. Akpe, A. R., Ekundayo, A. O., Aigere, S. P., & Okwu, G. I. Bacterial degradation of petroleum hydrocarbons in crude oil polluted soil amended with cassava peels. American Journal of Research Communication. 2015;3(7), 99-118.
27. Jessim, A. I., Hassan, R. K., & Salman, A. S. Isolation and identification of some pathogenic bacteria species from contaminated ambient with oily hydrocarbons. J Bacteriol Mycol Open Access. 2020;8(4), 111-115.
28. Kong, L., Gao, Y., Zhou, Q., Zhao, X., & Sun, Z. Biochar accelerates PAHs biodegradation in petroleum-polluted soil by biostimulation strategy. Journal of hazardous materials. 2018;343, 276-284.
29. Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S. & Bolton, E. E. PubChem in 2021: new data content and improved web interfaces. Nucleic acids research. 2021;49, 388-395.
30. Kuppusamy, S., Maddela, N. R., Megharaj, M., & Venkateswarlu, K. Fate of total petroleum hydrocarbons in the environment. Springer, Cham. Petroleum hydrocarbons. 2020;57-77.
31. Rehman, A. U., Nazir, S., Irshad, R., Tahir, K., ur Rehman, K., Islam, R. U., & Wahab, Z. Toxicity of heavy metals in plants and animals and their uptake by magnetic iron oxide nanoparticles. Journal of Molecular Liquids 2021;321-355.
32. Pérez‐Pantoja, D., Donoso, R., Agulló, L., Córdova, M., Seeger, M., Pieper, D. H., & González, B. Genomic analysis of the potential for aromatic compounds biodegradation in Burkholderiales. Environmental microbiology. 2012;14(5), 1091-1117
33. .Kumar, R. Pseudomonas Species: Natural Scavenger of Aromatic Compounds from Industrial Effluents. In Microbial Technology for the Welfare of Society, Springer, Singapore. 2019;273-287.
34. Edbeib, M. F., Wahab, R. A., Huyop, F. Z., Aksoy, H. M., & Kaya, Y. Further analysis of Burkholderia pseudomallei MF2 and identification of putative dehalogenase gene by PCR. Indonesian Journal of Chemistry. 2020;20(2), 386-394.
35. War Y, El-Hanafy A A, Sabir J S, Al-Garni S M, AlGhamdi K et al., Characterization of Mesophilic Bacteria degrading crude oil from different sites of Aramco, Saudi Arabia. Polycyclic Aromatic Compounds. 2017; 1-9.
36. Andhale JD, Misra RN, Gandham NR, Angadi KM, Jadhav SV, Vyawahare CR, et al. Incidence of Pseudomonas aeruginosa with special reference to drug resistance and biofilm formation from clinical samples in tertiary care hospital. J Pharm Biomed Sci. 2016;6:387–91.
37. Chikere, C. B., & Fenibo, E. O. Distribution of PAH-ring hydroxylating dioxygenase genes in bacteria isolated from two illegal oil refining sites in the Niger Delta, Nigeria. Scientific African. 2018;79(23), 7256-7263.
38. Hashim ST, Fakhry SS, Rasoul LM, Saleh TH, Alrubaii BA. Genotyping toxins of Clostridium perfringens strains of rabbit and other animal origins. Trop J Nat Prod Res. 2021;5(4):613-616.
39. Abdulkaliq Awadh H, Hammed ZN, Hamzah SS, Saleh TH, AL-Rubaii BA. Molecular identification of intracellular survival related Brucella melitensis virulence factors. Biomedicine. 2022; 42(4):761-765.
40. Abdul-Gani MN, Laftaah BA. Purification and characterization of chondroitinase ABC from Proteus vulgaris, an Iraqi clinically isolate. Curr Sci. 2017:2134-2140.
41. Kadhim AL-Imam MJ, AL-Rubaii BA. The influence of some amino acids, vitamins and anti-inflammatory drugs on activity of chondroitinase produced by Proteus vulgaris caused urinary tract infection. Iraqi J Sci. 2016; 57 (4A):2412-2421.
42. Saleh, T.; Hashim, S.; Malik, S.N.; Al-Rubaii, B.A.L. The impact some of nutrients on swarming phenomenon and detection the responsible gene RsbA in clinical isolates of Proteus mirabilis. Int J Pharm Sci Res. 2020;1(6):437-444.
43. Shehab ZH, Laftah BA. Correlation of nan1 (Neuraminidase) and production of some type III secretion system in clinical isolates of Pseudomonas aeruginosa. Biomed res. 2018;15(3):1729-1738.
44. Al-Rubii, BAL. Cloning LasB gene of Pseudomonas aeruginosa eiastase 10104-2aI in E. coli BL21 and E.coli DH5? and investigated their effect on the stripping of vero cells. Pakistan J Biotechnol.2017;14(4):697-705.
45. Abdulla L, Ismael MK, Salih TA, Malik SN, Al-Rubaii BA. Genotyping and evaluation of interleukin-10 and soluble HLA-G in abortion due to toxoplasmosis and HSV-2 infections. Ann Parasitol. 2022; 68(2):385–390.
46. Jiad AL, Ismael MK, Muhsin SS, Al-Rubaii BA. ND2 Gene Sequencing of Subfertile Patients Recovered from COVID-19 in Association with Toxoplasmosis. Bionatura. 2022; 7(3):45. http://dx.doi.org/10.21931/RB/2022.07.03.45.
47. Rasoul LM, Nsaif MM, Al-Tameemi MT, Al-Rubaii BA. Estimation of primer efficiency in multiplex PCR for detecting SARS-Cov-2 variants. Bionatura, 2022, 7(3): 48 / http://dx.doi.org/10.21931/RB/2022.07.
48. Rasoul, LM., Marhoon AA, Albaayit SFA, Ali RW, Saleh,TH, Al-Rubaii BAL. Cytotoxic effect of cloned EGFP gene on NCI-H727 cell line via genetically engineered gene transfer system. Biomedicine (India). 2022, 42(5): 938-942.
49. Ahmed AA, Khaleel KJ, Fadhel AA, Al-Rubaii BAL. Chronic Myeloid Leukemia: A retrospective study of clinical and pathological features. Bionatura, 2022, 7(3), 41. DOI. 10.21931/RB/2022.07.03.41.
50. Ali SM, Lafta BA, Al-Shammary AM, Salih HS. In vivo oncolytic activity of non-virulent Newcastle disease virus Iraqi strain against mouse mammary adenocarcinoma. AIP Conference Proceedings, 2021, 2372, 030010.
51. Ali SM, Laftah BA, Al-Shammary AM, Salih HS. Study the role of bacterial neuraminidase against adenocarcinoma cells in vivo. InAIP Conference Proceedings 2021, 2372, 030009.
52. Hamoode RH, Alkubaisy SA, Sattar DA, Hamzah SS, Saleh TH, Al-Rubaii BAL. Detection of anti-testicular antibodies among infertile males using indirect immunofluorescent technique. Biomedicine (India).2022; 42 (5):978-982.
53. Rasin KH, Algabar FA, Al-Obaidi M, Krayfa AH. EVALUATION OF MEDICAL WASTE MANAGEMENT DURING THE COVID-19 IN DIYALA GOVERNORATE. Environmental Engineering & Management Journal . 2022; 21(12):1931–1944.
54. Abbas MS, Ahmed AG, Ali SQ , AL-Rubaii BA. Immunological inflammatory factors in patients diagnosed with COVID-19. Biomedicine. 2023; 43(1):230-235.
55. Eyad HN, Adbulateed YA, Lafi SA. Abnormal presentation of TB patients: anthropological study, Ann Trop Med and Public Health. 2019; 22(VI): S186.
56. Abdulateef Y. Role of PSA in Diagnosis of Chronic Prostatitis. Indian Journal of Forensic Medicine & Toxicology. 2020;14(1):774-779.
57. Allami RH, Hassoon AH, Abdulateef YM, Ghani AA. Genetic Association of Angiotensin-converting enzyme 2 ACE-2 (rs2285666) Polymorphism with the Susceptibility of COVID-19 Disease in Iraqi Patients. Trop J Nat Prod Res. 2023;7(2):2346-2351.
58. Rasoul LM, Allami RH, Alshibib AL, laftaah Al-Rubaii BA, Sale TH. Expression and cytotoxic effect of recombinant Newcastle Disease Virus (rNDV) vector expressing enhanced green fluorescent gene in JHH5 cell line. Biomedicine. 2023; 43(1):205-209. doi: 10.51248/.v43i1.2425.
59. Jawad NK, Numan AT, Ahmed AG, Saleh TH, Al-Rubaii BA. IL-38 gene expression: A new player in Graves’ ophthalmopathy patients in Iraq. Biomedicine. 2023;43(1):210-215. doi: 10.51248/.v43i1.2027.
60. Bresam S, Al-Jumaily RM, Karim GF, Al-Rubaii BA. Polymorphism in SNP rs972283 of the KLF14 gene and genetic disposition to peptic ulcer. Biomedicine. 2023; 43(1):216-220. doi: 10.51248/.v43i1.2411
Received: 25 June 2023/ Accepted: 26 August 2023 / Published:15 September 2023
Citation: Fahim Muhsin K, AboKsour M F, Hadi S. Bioremediation by bacteria isolated from water contaminated with hydrocarbons. Revis Bionatura 2023;8 (3) 94 http://dx.doi.org/10.21931/RB/2023.08.03.94