2023.08.02.5
Files > Volume 8 > Vol 8 No 2 2023
Molecular identification of a Pseudomonas putida strain isolated from Machángara river (Quito-Ecuador) tolerant to carbamazepine
1 Universidad Politécnica Salesiana; Grupo de investigación BIOARN; [email protected]
2 Universidad Politécnica Salesiana; [email protected]
3 Universidad Politécnica Salesiana; Grupo de investigación GIDCARB; [email protected]
4 Universidad Politécnica Salesiana; [email protected]
* Correspondence: [email protected]; Tel.: (+593984794388, Quito-Ecuador)
Available from: http://dx.doi.org/10.21931/RB/2023.08.02.5
ABSTRACT
Carbamazepine is a molecule used to treat specific pathologies; however, it has become an emerging contaminant that is dangerous to the environment. In marine species and humans, it causes cytotoxicity, genotoxicity, reproductive disorders, and infertility. Thus, this pollutant has been subjected to conventional wastewater treatment, achieving low purification. In Ecuador, only some studies are related to emerging contaminants, and these show quantification but not treatment. Therefore, the implementation of biological techniques is necessary. In this sense, the research aimed to identify a bacteria in water samples from the Machangara River with carbamazepine tolerance. Morphological, biochemical, and molecular characteristics identified bacteria. The results indicated the presence of several microorganisms, including molecularly identified Pseudomonas putida. This was tolerant to carbamazepine concentrations of 15, 50, and 100 mg/L, with higher growth at the first concentration. This information can be valuable in wastewater treatment investigations.
Keywords: carbamazepine, degradation, Machangara river, Pseudomonas putida.
INTRODUCTION
Although water is an essential natural resource for life; however, it has been affected by natural and anthropogenic pollutants 1. During the development of humanity, these resources have been affected, and in recent years contamination has increased. In this sense, the term emerging pollutant appears, which includes some chemicals in low concentrations that alter the beneficial conditions of water, becoming toxic to the environment. Thus, primary investigation in emerging pollutant and biological treatments was shown in Europe and Asia and very little in Latin American countries such as Ecuador 2–4 . In turn, one problem that makes the study of emerging contaminants difficult is that there are limited techniques to identify low concentrations (ng/L to ug/L). In this sense, liquid chromatography becomes an alternative 3–5 .
On the other hand, carbamazepine is a drug used to treat epilepsy and trigeminal neuralgia. It is 72 % absorbed by the body and eliminated through the urinary and fecal tract 6. In this regard, the highest concentrations of carbamazepine have been found in water discharge from healthcare systems, either by the human body's elimination or by the mishandling of drugs 7 . The contamination is also due to an inefficient elimination in wastewater treatment plants 8 . Carbamazepine in the environment is associated with producing adverse teratogenic and mutagenic effects, reproductive problems, infertility, masculinization of females, feminization of males in animals, and genetic issues in human beings 2,9 . Hence, it is crucial to develop technologies for its removal.
In this regard, Reichert et al. 3 and Peña-Guzman et al. 4 pointed out that the concentrations of emerging contaminants detected in Latin America are often higher than in other countries; for example, in Ecuador, the authors Voloshenko et al.2 and Pacheco 10 identified a concentration between 1.5-830 µg/L of carbamazepine in Machangara, San Pedro, Guayllabamba, and Esmeralda rivers. Latin America does not have an established list of pollutants of concern or laws controlling their production or release to the environment 11 . Therefore, it is interesting to develop environment-friendly approaches to eliminate these contaminants; the first step is identifying microorganisms capable of tolerating them in high concentrations for future degradation tests.
MATERIALS AND METHODS
Reagents and standards
Chemical compounds studied in this article: Carbamazepine (≥ 99 % pure) was purchased from Sigma-Aldrich (Merck, Germany). Methanol and acetonitrile HPLC-grade were purchased from Dikma (DikmaTechnologies, USA).
Enrichment and isolation of bacteria potentially tolerant to carbamazepine
Isolation of carbamazepine-tolerant bacteria used the enrichment method. Thus, contaminated water was sampled, enriched with carbamazepine, and active bacteria that tolerated the contamination was isolated.
The contaminated water sample was obtained from the Machángara River in the Pichincha province in Quito-Ecuador (coordinates: 0°15'25.4"S 78°31'33.8"W) at 2644 masl. Thus, 250 mL of contaminated water was sampled in amber bottles, following the methodology of the sampling standard for microbiological analysis of the Ecuadorian Institute for Water Standardization (INEN) 12 . Sampling was performed in duplicate and at the same site.
Bacteria from the collected water samples were inoculated into a basal mineral medium (MBM). The medium used contains the following salts (g/L): K2 HPO4 3.5, KH2PO4 1.5, (NH4)2SO4 0.5, NaCl 0.5, MgSO47H2O 0.15, and 1mL of trace solutions. The trace elements contained: 2.0 g NAHC3-10H2O, 0.3g MnSO4-4H2O, 0.2g ZnSO4-7H2O, 0.02g (NH4)Mo7 O24 - 4H2O, 0.1g CuSO4 -5H2O, 0.5g CoCl2-6H2O, 0.05g CaCI2 -2H2O, 0.5g FeSO4 - 7H2O6 . Next, 15mg/L of carbamazepine was added. The culture was incubated at 26 °C, 150 rpm for 15 days in darkness to develop a community of microorganisms capable of producing the enzymes responsible for consuming this recalcitrant compound 6 . The bacteria were isolated using MBM with carbamazepine in a solid medium, and subcultures were carried out until obtaining pure cultures.
Bacterias tolerant to carbamazepine were identified, cultivating these in 250 mL of MBM with carbamazepine (15 mg/L, 50mg/L, and 100mg/L) as the only carbon source 6, as it is shown in Figure 1. Samples were incubated at 150 rpm in the orbital shaker (Shanghai, China) at 30 °C in darkness for 33 days. The tests used a bacterial inoculum concentration of 2% v/v. In turn, to monitor bacterial growth and carbamazepine consumption, samples were taken from the culture at the beginning and end of the study. The experiment used a control with MBM and carbamazepine without bacteria. Plate count was performed in TSA medium at 30 °C and expressed in colony forming units (CFUs) 13–15. All cultures were performed in triplicate.
Carbamazepine quantitation was performed by liquid chromatography using an HPLC Waters 1525 equipped with a diode-array detector and a binary pump, C18 column (Novapak® 4 x 300 mm). The HPLC conditions were as follows: a wavelength of 280 nm, a flow rate of 1 mL/min, an injection volume of 20 μl, and a retention time of 8.0 minutes. The mobile phase consisted of water: methanol: acetonitrile (50:25:25), pre-filtered by a 0.45 µm membrane and degassed 16 . A carbamazepine calibration curve was prepared with standards of 100 mg/L, 75 mg/L, 50 mg/L, 25 mg/L, 10 mg/L, and 5 mg/L. The chromatograms were monitored at 280 nm with the Empower™ 3 software (build 3471). The samples were analyzed in duplicate.
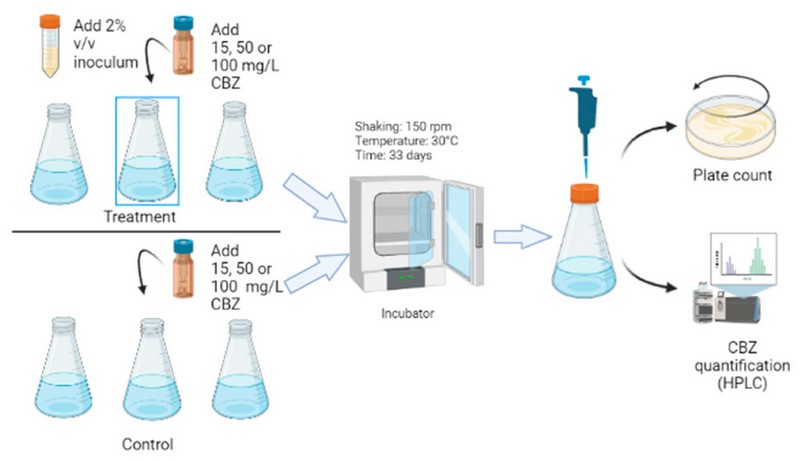
Figure 1. Tolerance essays include preparation of the inoculum, incubation, bacterial count, and quantification of carbamazepine (CBZ).
Morphological and biochemical characterization of the bacterial strain
The isolated bacteria strain was identified using morphological and biochemical techniques. Catalase and oxidase tests were used, and the Microgen TM GN-ID Identification Kit. The Microgen Identification Software (MID-60) was used to interpret the kit's results. Gram reactions were observed in cells cultivated for 24 h using an optical microscope 17 .
Molecular identification of the bacterial strain
The bacterium was cultured in test tubes with Tryptone Soy Agar for 24 h at 26 °C, and the DNA extraction process was performed with the Purelink Genomic DNA kit by Thermo Fisher Scientific 18 . DNA integrity was evidenced by electrophoresis with the 1KB Ladder Express DNA (molecular weight marker). The purity and concentration of the resulting DNA preparation were determined spectrophotometrically at 260 nm using a Quibit® 2.0 Fluorometer.
PCR targeted the 16s ribosomal genes (16s rRNA). First, the 16S rRNA gene was amplified using a forward primer 27F (5`-AGTTTGATCCTGGCTCA-3`) and a reverse primer 1492R (5`-GGTTACTCTTGTTACGACTT-3`) under the following conditions: 95 °C for 5 minutes, 40 cycles, 95 °C for 30 seconds, 56 °C for 30 seconds, 72 °C for 1 minute and final extension at 72 °C for 10 minutes 19 .
The success of amplification was checked in 1% agarose gel after amplification. The amplified fragment was sequenced by Molecular Research LP (MRDNA) in Texas-USA using an Illumina MiSeq platform following the manufacturer's guidelines (40 K average number of reads per assay). Sequence data were processed using the MR DNA analysis pipeline (MR DNA, Shallowater, TX, U.S.A.).
In summary, sequences <150 bp and ambiguous base calls were removed. Operational taxonomic units (OTUs) were generated, and chimeras were removed. OTUs were defined by 97% similarity. Final OTUs were taxonomically classified using BLASTn against the NCBI database. Sequencing reads from this study have been deposited in GeneBank with an accession number for the 16S rRNA gene (MW691110).
RESULTS AND DISCUSSION
Morphological and molecular characterization of the bacterial strain
Some bacteria were isolated from tolerance assays, and molecular identification was performed with one of them. The isolated bacterium showed a slightly yellow mucous colony with a smooth edge (Figure 2a). It was classified as a gram-negative bacillus (Figure 2b), oxidase and catalase positive 20–22 , typical characteristics of Pseudomonas 23,24 . Microgen identification Software (MID-60) identified the bacteria isolated as Pseudomonas pseudomallei with a 94.38% identity.
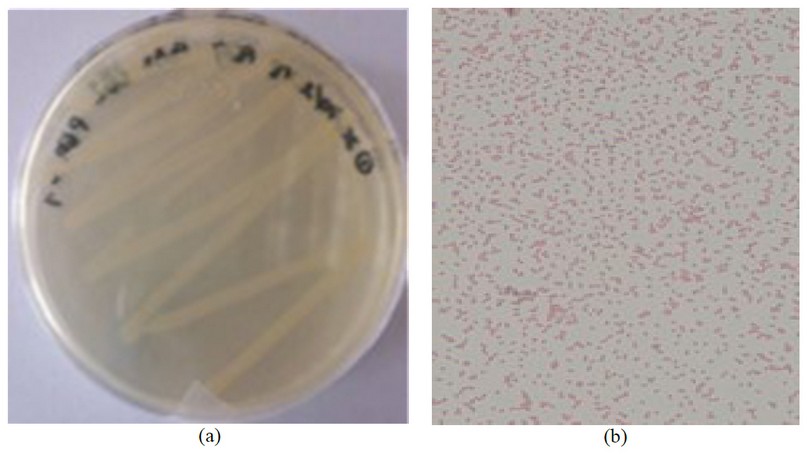
Figure 2. Bacterial identification. a) Isolated bacteria from Machángara river in TSA medium, b) Gram-negative Bacillus.
On the other hand, molecular identification by electrophoresis reported the presence of DNA, with a region size of approximately 1500 bp (Figure 3). These results suggest that this is the size corresponding to the 16s rRNA region, as established by other studies 25 . Finally, sequencing was performed with the amplified samples at the Molecular Research LP (MRDNA lab) laboratory in Texas-USA, obtaining a 100% identity of Pseudomonas putida for the strain.
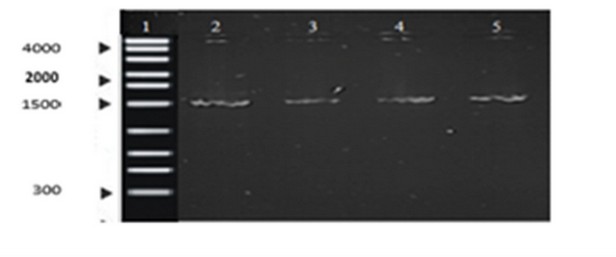
Figure 3. Agarose gel electrophoresis. Lane 1 represents the DNA ladder, and Lanes 2-5 show the PCR products from 4 samples of the strain in the study.
According to the literature, the bacteria identified as belonging to the Pseudomonadaceae family 26 , are used in degradation tests by having a metabolism that makes it easier to adapt to different habitats, sources of carbon, and nitrogen, that can colonize several ecological niches 27 . Therefore, these microorganisms, isolated from a contaminant-containing medium, are presumed to tolerate toxic substances and may help degrade carbamazepine 28 . Pseudomonas putida is not associated with human disease 29 , offering many advantages such as cost-effectiveness and utilization of native harmless microbial species 30
Carbamazepine tolerance assays
Pseudomonas putida reached the highest biomass concentration at 15mg/L with a carbamazepine consumption of 49.56% (SD:5.6), which indicates that bacteria use the contaminant as a carbon source because there was no reduction of CBZ in controls. Bacterias showed poor growth at 50 and 100 mg/L (Figure 4), with 11.3% (SD:2) and 9.9% (SD:3.2) of degradation, respectively.
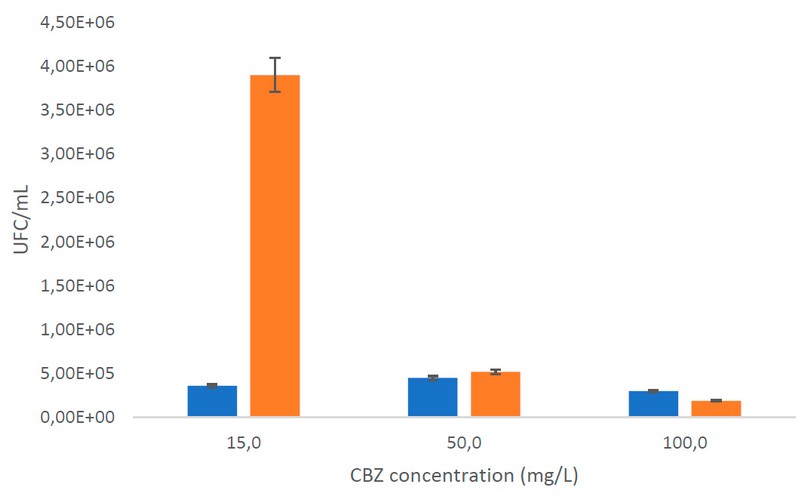
Figure 4. Plate count. The orange bar is the initial UFC/mL; the blue bar is the final UFC/mL
Figure 4 shows that bacterial growth and degradation were low at higher concentrations. Thus, the decrease in microorganisms could be because the bacteria finished the exponential phase, and the death phase began 31. In addition, it should be considered that there are several kinetic models in biodegradation and that, in some instances, the concentration of organisms remains constant even when the substrate is degraded by inhibition. Therefore, it is associated with a decrease in specific growth rates. For example, in the study of Rathour et al. 32 , it was shown that the specific growth rate was rapidly reduced due to the detrimental effects of benzo(a)pyrene on cell development and the subsequent inhibition of the substrate on culture growth. Likewise, some species of Pseudomonas spp use bioabsorption as a bioremediation mechanism that causes the accumulation of toxic compounds inside the cell, causing its death 33 .
In the study conducted by (Li et al. 34 , similar carbamazepine consumption percentages were observed, with values of 46.6%, where the drug was observed to decrease at day 6 (144 hours) with Pseudomonas sp. CBZ-4. In the study by Ha, Mahanty, Yoon, & Kim 35 , with mixed bacterial culture (Pseudomonas, Bacillus, and Acinetobacter), a 60% degradation was obtained in 12 days. It should also be noted that several studies have determined that microorganisms require a long period of prior adaptation to persistent chemical contaminants to degrade them effectively 36 . In this study, the strains evaluated were acclimated for 15 days to increase their tolerance.
Figure 4 shows that Pseudomonas putida has a better tolerance for low concentrations of carbamazepine, like the study of Gheorghe et al. 37, considering that pharmaceutical biodegradation could be affected by different factors such as temperature, pH, varying pharmaceutical concentrations, etc. For this reason, it is necessary to carry out future degradation tests with different culture conditions to determine the optimal ones to apply in wastewater treatment. According to the literature, carbamazepine, naproxen, and ibuprofen have harmful effects at 16.21 to 54.21 mg/L in bacteria, indicating that they influence the degradation route of the microorganism at higher concentrations, causing toxicity and inhibiting its metabolization. It is confirmed by Nogales et al. 38 , who indicate that contaminants at high concentrations cause severe damage to the bacterial cell, causing death 38 . Similar results with emerging contaminants were mentioned by Ramos et al. 39 for Pseudomonas putida in degradation tests of aromatic hydrocarbons and short-chain alcohols; also, the degradation was reported of salicylic acid by P. fluorescens HK44 was inhibited at high concentrations of the substrate 40 . However, the concentration of carbamazepine in wastewater is usually found at concentrations below that evaluated in this study 2 .
The capacity of Pseudomonas putida to tolerate carbamazepine could be due to the presence of central pathways to degrade these compounds, and a multifactorial response is generated to adapt to the environment when exposed to these, involving degradation routes and metabolism changes as a result of stress 38 . Pseudomonas putida produces oxidoreductases, hydrolases, and thiolases that help the microorganism overcome the damage caused by pollutants 39 . In addition, this bacterium finds new catabolic pathways to metabolize organic compounds 41 by presenting the catabolic TOL pWW0 plasmid, which, according to the literature, encodes specific metabolic activities so that the bacterium grows and uses it as the only source of carbon and energy, being closely related to biodegradation applications 42 .
CONCLUSIONS
Carbamazepine is an emerging pollutant in the environment, resistant to conventional degradation processes, so microorganisms may be an option to biodegrade. Pseudomonas putida was molecularly identified from the wastewater from the Machángara River when exposed to 15, 50, and 100 mg/L of the contaminant carbamazepine at 30 °C temperature and evaluated over 33 days. Pseudomonas putida used carbamazepine as the sole carbon source during in-vitro tolerance assays, resulting in a reduction of 49.56 % at 15 mg/L of the contaminant quantified by HPLC. In conclusion, these microorganisms could be used in future degradation studies as a sustainable technology solution to be developed and applied for the recovery of contaminated environments, considering the ability of P. putida to increase in several hostile environments. In addition, bacteria isolated from a natural source exposed to this contaminant could be advantageous in bioremediation since biodegradation rates depend on different factors, such as previous adaptation of the microbial inoculum.
Author Contributions: methodology, Méndez Gabriela, Morales Leslie, Garzón Valeria.; formal analysis, Méndez Gabriela, Morales Leslie, Coyago Elena.; investigation, Méndez Gabriela, Morales Leslie, Garzon Valeria, Coyago Elena.; resources, Méndez Gabriela.; writing—original draft preparation, Méndez Gabriela, Morales Leslie, Coyago Elena.; writing—review and editing, Méndez Gabriela, Morales Leslie, Garzon Valeria and Coyago Elena; project administration, Méndez Gabriela.; funding acquisition, Méndez Gabriela. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement: Sequencing reads from this study have been deposited in GeneBank with accession numbers for 16S rRNA gene MW691110 (https://www.ncbi.nlm.nih.gov/nuccore/MW691110)
Conflicts of Interest: "The authors declare no conflict of interest."
REFERENCES
1. Franco, P. & Rodríguez, M. Formulación de líneas estratégicas para un proyecto participativo de conservación de un ojo de agua de la parroqui El Condado, barrio Colinas del norte, sector El manatial y sector Rancho bajo. Pontifica Universidad Católica del Ecuador (Pontificia Universidad Católica del Ecuador, 2016).
2. Voloshenko-Rossin, A. et al. Emerging pollutants in the Esmeraldas watershed in Ecuador: discharge and attenuation of emerging organic pollutants along the San Pedro–Guayllabamba–Esmeraldas rivers. Environ. Sci.: Processes Impacts 17, 41–53 (2015).
3. Reichert, G., Hilgert, S., Fuchs, S. & Azevedo, J. C. R. Emerging contaminants and antibiotic resistance in the different environmental matrices of Latin America. Environmental Pollution 255, (2019).
4. Peña-Guzmán, C. et al. Emerging pollutants in the urban water cycle in Latin America: A review of the current literature. J Environ Manage 237, 408–423 (2019).
5. Varo, P., López, C., Cases, V. & Ramírez, M. Presencia de contaminantes emergentes en aguas naturales. 33 Preprint at (2016).
6. Gauthier, H., Cooper, D. G. & Yargeau, V. Biodegradation of pharmaceuticals by common microorganisms. in WIT Transactions on Ecology and the Environment vol. 111 263–271 (2008).
7. Rojo, M. et al. Human pharmaceuticals in three major fish species from the Uruguay River (South America) with different feeding habits. Environmental Pollution 252, 146–154 (2019).
8. Viancelli, A. et al. A review on alternative bioprocesses for removal of emerging contaminants. Bioprocess Biosyst Eng 43, 2117–2129 (2020).
9. Robledo, Z. et al. Hidroquìmica y contaminantes emergentes en aguas residuales urbano industriales de Morelia, Michoacàn, México. Revista Internacional de Contaminacion Ambiental 33, 221–235 (2017).
10. Pacheco, A. Determinación de contaminantes emergentes carbamazepina y diclofenaco en el Río San Pedro y análisis de tratamiento con nanotecnología. (Universidad de las Fuerzas Armadas del Ecuador, 2021).
11. Zhang, Y., Geißen, S. U. & Gal, C. Carbamazepine and diclofenac: Removal in wastewater treatment plants and occurrence in water bodies. Chemosphere 73, 1151–1161 (2008).
12. INEN. Aguas. Muestreo para examen microbiològico. Norma Técnica Ecuatoriana (2012).
13. Camacho, A. et al. Cuenta en placa de bacterias. in (2009).
14. Sánchez, E. P., Núñez, D., Cruz, R. O., Torres, M. A. & Herrera, E. V. Simulación y Conteo de Unidades Formadoras de Colonias Simulation and Counting of Colony-Forming Units. vol. 28 (2016).
15. Matheus, P., Ramírez, E., Araque, J., Belandria, O. & Durán, J. Espectrofotometría de absorciòn molecular ultravioleta como una herramienta para estudiar el crecimiento de las cepas ATCC 25922 y ATCC 35218 de Escherichia coli. (2016).
16. Garzón, G. & Popayán, M. Estandarización de un método analítico por cromatografía líquida de alta eficiencia para la cuantificación de carbamazepina en tabletas. 130–137 (2008).
17. López-Jácome, L. E. et al. Las tinciones básicas en el laboratorio de microbiología. Investigación en Discapacidad 3, 10–18 (2014).
18. Life. PureLink ® Genomic DNA Kits For purification of genomic DNA.
19. Chen, Y. et al. Obtaining long 16S rDNA sequences using multiple primers and its application on dioxin- containing samples. 16, 1–11 (2015).
20. Pírez, M. & Mota, M. Morfología y estructura bacteriana. in 23–42 (2008).
21. Winn, W. C. & Koneman, E. W. Koneman diagnóstico microbiológico : texto y atlas en color. (Editorial Médica Panamericana, 2008).
22. Cuesta, C. & Pérez, J. Presencia de cepas de Staphylococcus spp. meticilino resistentes con resistencia inducible a la clindamicina de pacientes hospitalizados en el “Hospital Vicente Corral Moscoso (HVCM)”. (Universidad de Cuenca, 2016).
23. Bou, G., Fernández-Olmos, A., García, C., Sáez-Nieto, J. A. & Valdezate, S. Métodos de identificación bacteriana en el laboratorio de microbiología. Enfermedades Infecciosas y Microbiologia Clinica vol. 29 601–608 Preprint at https://doi.org/10.1016/j.eimc.2011.03.012 (2011).
24. Rojas-Triviño, A. Conceptos y prácticas de microbiologia general. (2011).
25. Ortíz, Á., Martínez, M. & Vargas, F. La secuencia completa del gen ARN ribosomal 16s, una promesa para mejorar laa presión en la asignación taxonómica microbiana. Microbiología Ambiental en México 22 (2017) doi:978-607-02-9617-8.
26. Molina, J. & Orozco, J. Detección de resistencia antimicrobiana en bacterias de interés clínico aisladas en el Río Chambo. Noviembre 2018 - enero 2019. (Universidad Nacional de Chimborazo, 2019).
27. Estupiñán, M. et al. Aislamiento e identificación de Pseudomonas sp. y Aeromonas sp. en aguas de piscinas públicas de Bogotá – Colombia. NOVA 25–29 (2017).
28. Ríos, S., Agudelo, R. & Gutiérrez, L. Patógenos e indicadores microbiológicos de calidad del agua para consumo humano. (2017) doi:10.17533/udea.rfnsp.v35n2a08.
29. Goverment of Canada. Pseudomonas putida - information sheet - Canada.ca. https://www.canada.ca/en/health-canada/services/chemical-substances/fact-sheets/chemicals-glance/pseudomonas-putida.html (2017).
30. Wang, S. N. et al. Biodegradation and detoxification of nicotine in tobacco solid waste by a Pseudomonas sp. Biotechnol Lett 26, 1493–1496 (2004).
31. Kareem, S., Kefas, H., Chior, T. & Latinwo, G. View of Kinetics of Fermentation by Enzymes: A Mathematical Approach. AU Journal of Technology 15, 109–114 (2011).
32. Rathour, R. K., Sharma, D., Sharma, N., Bhatt, A. K. & Singh, S. P. Chapter 14 - Engineered microorganisms for bioremediation. in Current Developments in Biotechnology and Bioengineering (eds. Joshi, S., Pandey, A., Sirohi, R. & Park, S. H.) 335–361 (Elsevier, 2022). doi:https://doi.org/10.1016/B978-0-323-88504-1.00002-9.
33. Vélez, J. M. B., Martínez, J. G., Ospina, J. T. & Agudelo, S. O. Bioremediation potential of Pseudomonas genus isolates from residual water, capable of tolerating lead through mechanisms of exopolysaccharide production and biosorption. Biotechnology Reports 32, e00685 (2021).
34. Li, A. et al. Characterization and biodegradation kinetics of a new cold-adapted carbamazepine-degrading bacterium, Pseudomonas sp. CBZ-4. Journal of Environmental Sciences 25, 2281–2290 (2013).
35. Ha, H., Mahanty, B., Yoon, S. & Kim, C.-G. Degradation of the long-resistant pharmaceutical compounds carbamazepine and diatrizoate using mixed microbial culture. Journal of Environmental Science and Health, Part A 51, 467–471 (2016).
36. Poursat, B. et al. Does long term exposure leads to biodegradation of carbamazepine ? (2016).
37. Gheorghe, S., Petre, J., Lucaciu, I., Stoica, C. & Nita, M. Risk screening of pharmaceutical compounds in Romanian aquatic environment. Environ Monit Assess (2016) doi:10.1007/s10661-016-5375-3.
38. Nogales, J., García, J. & Díaz, E. Degradation of Aromatic Compounds in Pseudomonas : A Systems Biology View. (2017) doi:10.1007/978-3-319-39782-5.
39. Ramos, J. L. et al. Mechanisms of solvent resistance mediated by interplay of cellular factors in Pseudomonas putida. FEMS Microbiol Rev 39, 555–566 (2015).
40. Silva, T. R., Valdman, E., Valdman, B. & Leite, S. G. F. Salicylic acid degradation from aqueus solutions using Pseudomonas fluorescens HK44: parameters studies and application tools. Brazilian Journal of Microbiology 38, 39–44 (2007).
41. Lacal, J. Caracterización bioquímica y moloecular del sistema de dos componentes TODS/TODT de Pseudomonas putida DOT-T1E. (Universidad de Granada, 2008). doi:9788433849175.
42. Nikel, P. & de Lorenzo, V. Pseudomonas putida as a functional chassis for industrial biocatalysis: From native biochemistry to trans-metabolism. Metab Eng 50, 142–155 (2018).
Received: 26 December 2022 / Accepted: 15 March 2023 / Published:15 June 2023
Citation: Méndez G.; Coyago-Cruz, E.;Morales, L.; Garzón, V. Molecular identification of a Pseudomonas putida strain isolated from Machángara river (Qui-to-Ecuador) tolerant to carbamazepine. Revis Bionatura 2023;8 (2) 5. http://dx.doi.org/10.21931/RB/2023.08.02.5
