2023.08.02.13
Files > Volume 8 > Vol 8 No 2 2023
Production of nanobodies in Andean camelids and their most common applications: A general review in the medical field.
1Departamento de Ciencias de la Vida y la Agricultura, Laboratorio Multidisciplinario, Universidad de las Fuerzas Armadas – ESPE, Sangolquí, Ecuador.
2Centro de Nanociencia y Nanotecnología – CENCINAT, Universidad de las Fuerzas Armadas ESPE, Sangolquí, Ecuador
3Universidad Técnica de Machala, UTEMACH Machala, Ecuador
*Corresponding author. [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.02.13
ABSTRACT
The heavy chain fraction present in Camelidae antibodies is so-called nanobodies. They have different characteristics when compared to immunoglobulin G, like more diminutive size, higher affinity, shorter half-life in serum, etc. These proteins are codified by B lymphocytes cDNAs and can be produced in different hosts like Escherichia Coli, Pichia Pastoris, plant cells and even insect cells. Andean camelids have been mainly used in the Andean region of South America as transport means and source of raw materials like fibers and meat, then being of great economic importance. However, in Ecuador, the potential of these animals as a source of biomedical products has not been investigated or exploited yet. Due to the scarce information related to these molecules and their industrial production in the country, this review aims to remark on the most common medical application of nanobodies produced from Andean camelids; also, industrial applications are described.
Keywords: Cancer, Coronavirus, VHH, production, treatment, diagnosis.
INTRODUCTION
Camelid produces antibodies composed of heavy chain constant domains and a single variable domain; this variable domain is known as nanobody (Nb). It possesses a single antigen-binding domain called VHH 1,2. Antibodies with a single variable chain, known as vNARs, have also been reported in the fish subclass Elasmobranchii to which sharks and rays belong 3–5. Phylogenetic studies have shown that antibodies from Camelidae and Elasmobranchii evolved independently 6,7. In recent years, Nb has been used as therapeutic agents, in diagnostic tests and as research tools 8–10. Camelidae and Elasmobranchii possess different kinds of antibodies6, but only the antibodies that lack the light chains are of interest in this review.
Camelidae is a family that includes the old-world camelids: -Camelus dromedarius (dromedaries), and C. bactrianus (camels)- and the new-world camelids 11–13. There are four species of new-world camelids in South America 14,15: Lama glama (llamas), L. guanicoe (guanacos), Vicugna vicugna (vicuñas) and V. pacos (alpacas). All of them represent a genetic resource of great importance, not only from a scientific point of view 16–18, since they could represent an opportunity for economic, social and technological development in Ecuador.
Nbs were first discovered and described by the Hamers-Casterman team 19 at the Free University of Brussels while analyzing C. dromedarius serum. The potential use of Nbs in the medical field is focused on the prevention, diagnosis and disease treatments 20,21. Additionally, several preclinical and clinical studies regarding their use in phases 1 and 2 22,23. The development of a drug at the laboratory scale until its sanitary registration takes about 10 years24–26. In fact, in 2019, the Food and Drug Administration (FDA) approved for the first time the therapeutic use of a humanized Nb 27. Due to the great potential of Nbs, Steeland et al. 28 concluded that these molecules could be positioned as pharmaceuticals in a short period for daily use.
The main objective of this review is to describe the most common medical application of nanobodies produced from Andean camelids to demonstrate the potential of these molecules as a medical product. In addition, a little overview of its recombinant production in different biotechnological hosts and other industrial fields is described.
Immunoglobulin G VS Nanobodies
Structurally, immunoglobulins G possess 2 light and 4 heavy chains. Heavy chains contain 3 constant domains and 1 variable domain (VH), while the light chains contain 1 constant and 1 variable domain (VL) 29,30. However, Camelidae antibodies lack the light chain 31, and their 3 chains are composed of 2 constant domains and 1 variable domain (Nb) 32,33. In addition, Camelidae-like shark antibodies (which lack a light chain and possess 7 chains) are called New Antigen Receptors (NAR) antibodies 34. They comprise 5 constant domains and 1 variable domain (vNAR) 6,35. Figure 1 shows the structural comparison between mammalian IgG and the IgG-like antibodies from Elasmobranchii and Camelidae. Figure 2 shows the alignment of Elasmobranchii vNAR, Camelidae Nb and a Mus musculus heavy chain antibody sequences.
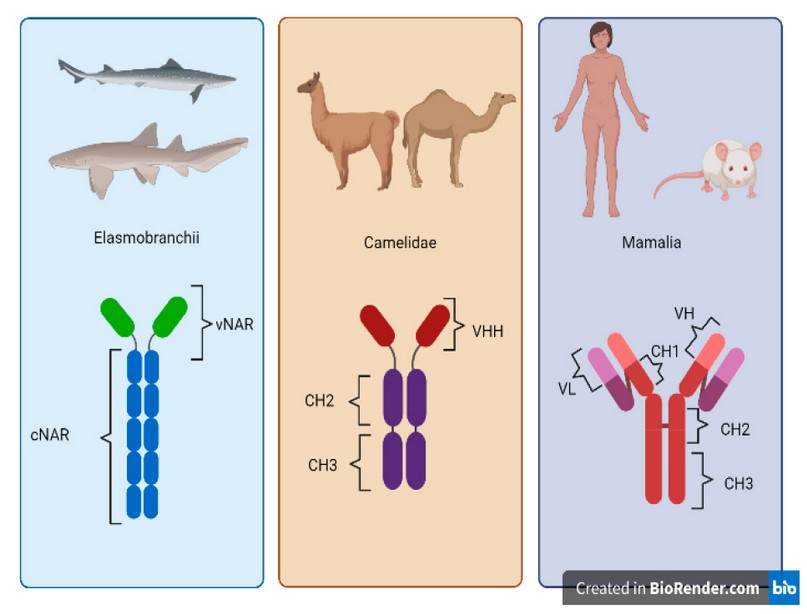
Figure 1. Structural comparison between IgG-like Elasmobranchii, Camelidae and Mammalian antibodies. Elasmobranchii antibodies possess a variable region called vNAR and a constant region composed of five constant domains (cNAR). Camelidae antibodies are conformed by one variable heavy chain domain (VHH) and 2 constant heavy chain domains (CH2 and CH3). Mammalian antibodies are composed of one variable heavy chain domain (VH), three constant heavy chain domains (CH1, CH2, CH3) and a constant light chain domain (VL).

Figure 2. Sequence logo plot of the alignment of vNAR (1T6V), VHH (1MEL) and monoclonal Mus musculus heavy chain (1MLC).
The figure shows the amino acid variation between vNAR, VHH and mAb heavy chains. In parenthesis protein name according to DATA bank information.
Despite the biotechnological importance of immunoglobulin G, Nbs have certain differences over IgG, which are summarized in Table 1.
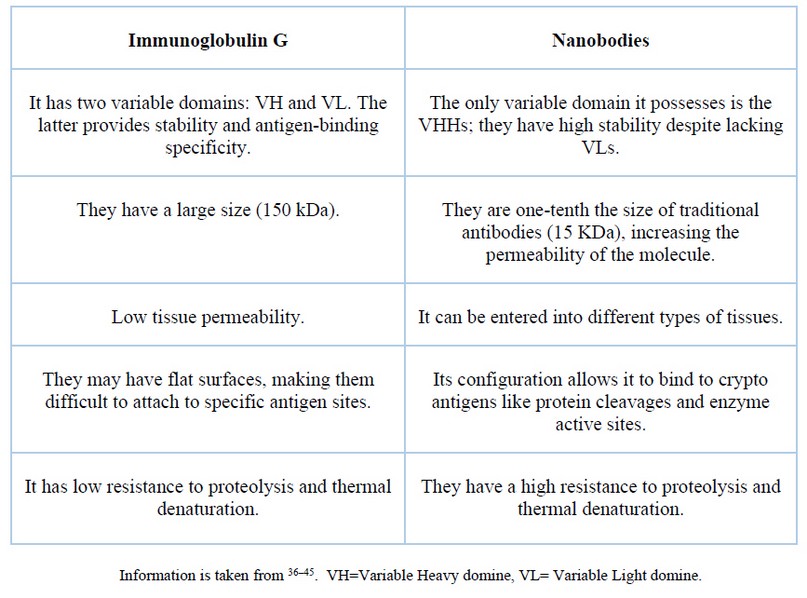
Table 1. Comparison between conventional antibodies and nanobodies.
The main Nbs disadvantage is their low half-life in human blood serum because they are filtered by the kidney in 26 to 60 minutes46–48. However, they have low toxicity and can be eliminated from the human body by filtration by the kidneys 49. Nbs have a low immunogenicity risk profile, which drives their use to develop potential clinical applications 50. Using Nbs conjugated with small molecules like drugs, toxins, enzymes and imaging agents allows an approach to target sites with low systemic toxicity 51. In addition, Nbs do not present the solubility and aggregation problems typical of conventional antibodies due to their hydrophilicity 36,52.
The size of a camelid antibody is around 95 kDA 53–55 due to the absence of the light chain; Elasmobranchii antibodies size is about 175 kDA56 while the variable antigen-binding domains (Nbs and vNAR) measure between 12 and 15 kDA 46,56,57. There is a vast difference between the IgG medical development and the research performed on Nbs and vNAR molecules 58. Nevertheless, Nb and vNAR have similar advantages to the IgG (more specificity, smaller capacity to bind crypto antigens, more stability) 59,60. Figure 3 represents the 3D structure of a Nb, a vNAR and a Mus musculus monoclonal antibody (mAb). This figure shows that the Nb and vNAR structures are smaller and more straightforward than the M. musculus mAb, facilitating their production 58.

Figure 3. 3D structure of antibody domains. a) VHH (1MEL). b) mAb of Mus musculus (1MLC). c) cNAR (1T6V). Information and images are taken from 61–63.
How are Nanobodies produced?
The development of Nb libraries can be achieved from a naive or synthetic source or by immunization. All of them are great ways to get the appropriate Nb that best meets the final research goal. For this reason, Nbs are expressed and produced at a large scale in the chosen host 64. Libraries obtained by immunization are the most widely used and consist in injecting the animal with the correct antigen 65. Native libraries are developed from non-immunized camelids blood 66, and synthetic libraries are developed from a determined sequence framework 67,68 or the target antigen 69, from which randomization of oligonucleotides in the hypervariable regions is performed 70. The yields in the production of Nbs from immune libraries and synthetic libraries can vary significantly71.
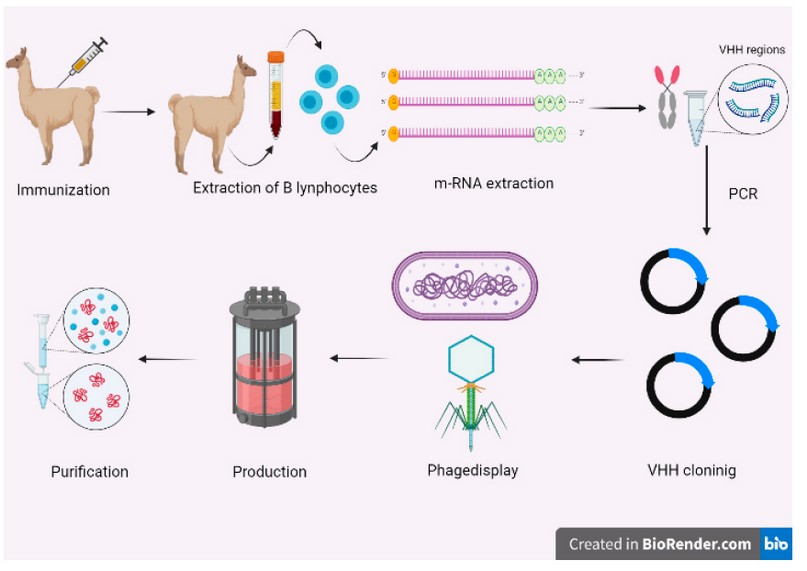
Figure 4. Production process of recombinant nanobodies. The first step is to immunize the camelide. Then, the B lymphocytes must be extracted and purified to extract m-RNA. The VHH sequences are obtained by PCR. Thereafter, the nanobody sequences are selected by phage display (or any display method). Finally, the chosen nanobodies are produced recombinantly and purified.
Figure 1 shows a general diagram for Nb production by immunization. This process starts with the inoculation of a young-adult camelid with 50 to 200 𝜇g of the chosen antigen 72. After the first immunization, the animal must be injected from 4 to 8 times during 2 months 64. Once the immunization process is completed, the camelid's blood must be extracted and B lymphocytes purified. Further, B lymphocyte mRNA is isolated, purified and recovered to finally get the total complementary cDNA using polymerase chain reaction (PCR) techniques 73,74.
Then, a Nb library is generated from the above-obtained cDNA, for which the VHH regions are amplified using specific primers 73,75. Muyldermans (2021) research suggests that the amplification of the VHH regions should be performed using mainly the nested PCR technique 64.
From the PCR products, a gene library is generated 67; then the fragments must be chosen by a display technique; the most common technique is the phage display 64. The phage display library consists of bacteriophages with the antigenic molecule (Nb) in their coat; then, all the library is exposed to the specific antigen 76. Finally, the selected fragments are screened and then chosen those that produce specific Nbs for the particular antigen 64,73,75,77.
The selected VHH sequences are cloned at the industrial level in appropriate hosts like bacteria such as Escherichia coli 64,78,79 or yeasts such as Pichia pastoris 75. Mammalian cells 73, insects or plants 71 are also excellent options as industrial production hosts 80. Table 2 exposes the kind of expression and yields of the different host cells. Finally, the proteins are extracted and purified, generally using the ammonium sulfate precipitation method in conjunction with other types of chromatography 73.
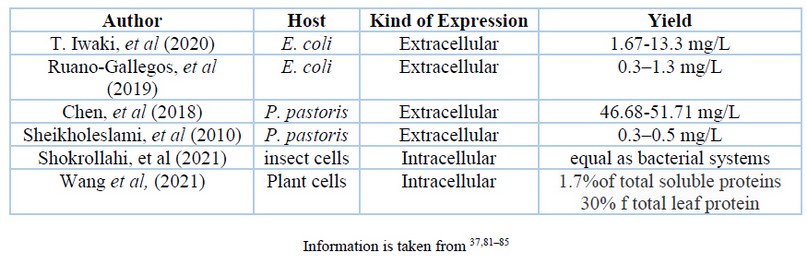
Table 2. Comparison between conventional antibodies and nanobodies.
Applications of camelid nanobodies in Human Health
The small Nb size, joined with their high stability, solubility and a great capacity to recognize crypto-antigens place them as excellent alternatives for the prevention 86, diagnosis 87,88 and human diseases treatment 46,89,90.
Disease diagnosis using Nbs points to:1- the generation of images from tissues and organs 57,91; and 2- the performance of immunoassays to detect pathogen antigens 89,92,93. Meanwhile, nanobodies as a therapeutic agents seek to attack an antigen, mainly to block virus replication 94,95 or bacteria growth 96.
Another novel application is their use in antiphonic serums as neutralizers and blockers of toxins present in different types of poisons 97–99.
Finally, a search was made for medical applications of nanobodies developed in Andean camelids. About 518 articles were found using the "Publish or perish" program where Andean camelids were used. The reported applications belonged mainly to the health branch and articles published in the last 5 years including 50 scientific publications that were not review articles.
Using descriptive statistics and frequency histograms, it was determined that the most common application of Nbs is the treatment and diagnosis of infectious diseases (Figure 3). However, the condition for which the largest number of Nbs have been developed was cancer (Figure 4).
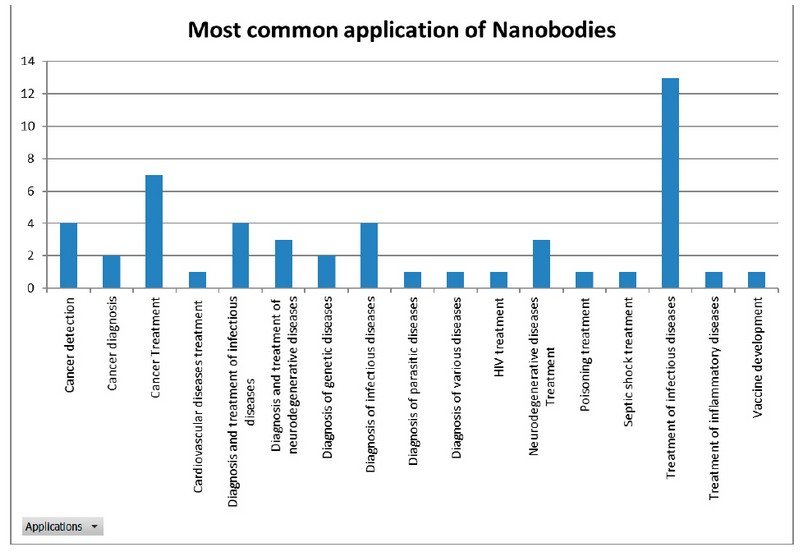
Figure 5. Most common applications of nanobodies.
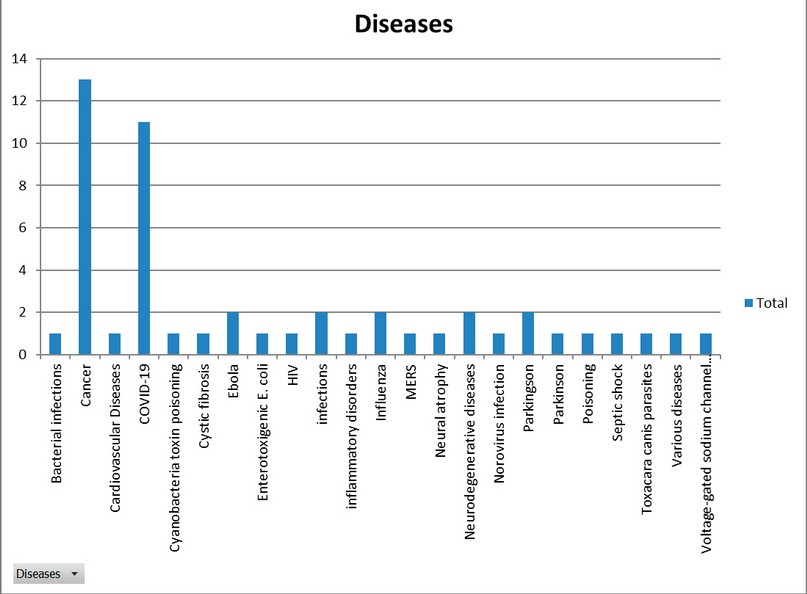
Figure 6. Diseases for which nanobodies were developed.
The 2020 pandemic greatly spurred the development of Nbs and other pharmaceuticals for the treatment and/or diagnosis of COVID-19 100,101. However, Nbs development focused on diagnosis, as shown in table 3; cancer treatment has been developed since before 2020.
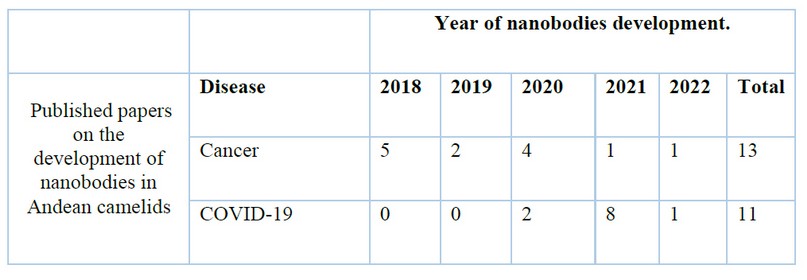
Table 3. Comparison between published papers reporting the development of Nanobodies in Andean camelids for cancer and COVID-19.
Cancer is a disease in which uncontrolled cell multiplication can spread to other parts of the body 102. Alternatives for cancer prevention have existed since the 1700s 103, and their treatments include surgeries, chemotherapies, radiotherapies, immunotherapies and hormone therapies 104. Nbs have a good synergy with these treatments105, so they could be used together, although the full range of Nbs applications in cancer has not yet been explored thoroughly 106,107.
On the other hand, COVID-19 caused by SARS-CoV-2 generates affections on the respiratory system 108. On March 11 of 2020, the World Health Organization (WHO) declared a world pandemic for this infection 109. There is still no defined treatment for this disease, although monoclonal antibodies 100,110 and nanobodies seem to be the best option to fight the virus 111–113. The future evolution of the virus must be awaited as it could generate less infectious and virulent variants or more aggressive strains resistant to the current preventive treatments 114.
Applications of camelid nanobodies in different branches of industry
The main focuses of Nbs in plants are to protect crops by providing immunity against pathogens 72,85 and rapid detection of toxins that may affect food safety 115. Passive immunity is the use of external antibodies to protect a patient 116–118; this concept can be extrapolated to plants, where antibodies would protect them against pathogens 85,119,120. On the other hand, Nbs can identify toxins in absorbance, fluorescence or Enzyme-Linked ImmunoSorbent Assay (ELISA) tests, depending on the toxin to be tested 121–123.
In the livestock sector, the main Nbs application is on early disease detection 124,125 to avoid zoonoses 3,126,127. Nbs can also provide passive immunity in animals, especially those sensitive to diseases after weaning, such as piglets 128. This is very important to avoid using antibiotics that could generate resistant bacterial strains 89,129.
Nanobodies used in water and soil allow the quick and accurate identification and detection of toxins and contaminants to the nanogram levels so, becoming a handy tool for these purposes 130–132.
The ability of nanobodies to bind to specific compounds makes them good alternatives in high-affinity chromatography 133,134. They can also help to determine the protein structure 135 with dynamic properties in case of proteins with various conformations and shapes which are difficult to solubilize 64,136. Another application is identifying protein functions and protein-protein interactions by mass spectrometry that could replace conventional antibodies by Nbs 137.
The use of intrabodies that consist of nucleic acid sequences with the genetic coding information of an antibody or Nb 138,139 that can be expressed inside a cell to modify cellular activity 140,141. This biological application could be used to alter cellular functions 142, blocking or reducing the activity of endogenous proteins 143.
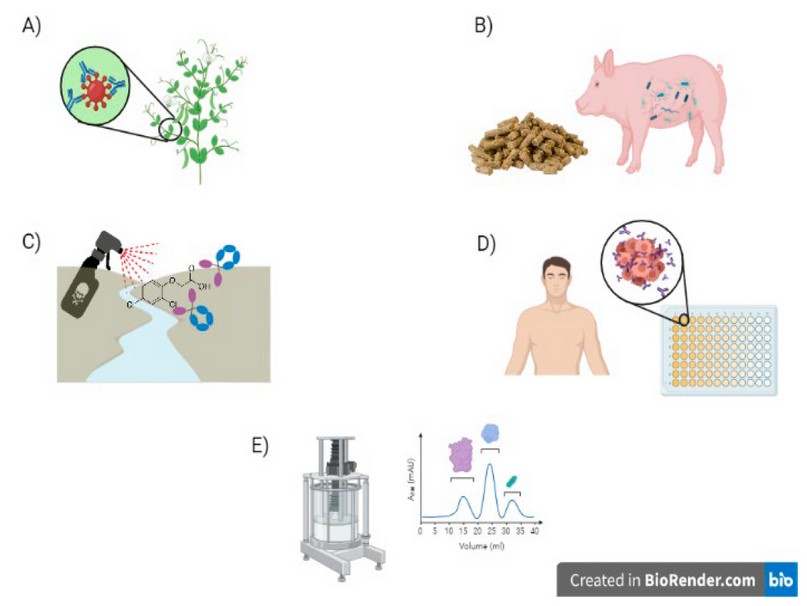
Figure 7. Applications of Nanobodies (Nb). A) Passive immunity in plants. B) Passive immunity in animals. C) Identification of contaminants in soil and water, D) Identification of human diseases. E) Nbs as a solid phase in chromatography.
Future Perspectives and Conclusions
This mini-review presented applications of nanobodies that could promote the use of Andean camelids in Ecuador as new sources of products of high social, economic and public health interest.
The SARS-CoV-2 virus generated a global health crisis. Therefore, several methods for the diagnosis of COVID-19 were rapidly developed 144. Nonetheless, there is still no defined treatment 100,110. Becker's study in 2020 145e mentions that a great deal of research is being carried out, mainly focused on blocking the replication and translation of the virus RNA; moreover, some treatments seek to block the binding of the virus and its ACE2 receptors 146–148 so, Nbs could be used as therapeutic molecules against COVID-19.
Ecuador has the greatest number of reptile species per unit area; one of the groups of greatest interest are snakes. Among them, about 35 species are reported to be venomous and are distributed along the Ecuadorian coast and Amazon 149. On the other hand, scorpions, of which 40 species are known to be venomous, have been little studied 150. In the past, serums to treat poisonings were produced by the "Instituto Nacional de Investigación en Salud Pública" INSPI, but production was suspended in 2014 so only antivenoms were imported 151. In 2022, INSPI produced a batch of 300 experimental antivenom sera 152. The Nbs could be used to act as high-affinity antivenom.
The incidence of cancer in Ecuador is 61% in women while 41% in men 153; the types of cancer most reported in male patients are prostate, lymphoma and stomach 154, while for female patients, they are breast cancer, cervix and lymphoid leukemia 153. Nanobodies could be an interesting alternative for treating these types of cancer in Ecuador. However, the fact that they are rapidly filtered by the kidney makes them less attractive as a therapeutic alternative 58.
In conclusion, the most common application of Nbs after analyzing more than 50 articles was the treatment and diagnosis of infectious diseases and cancer, so the future development of nanobodies should be linked to the treatment and diagnosis of these diseases. It is suggested that Nbs that achieve a balance between tissue permeability and a longer half-life in serum be developed for treating diseases such as cancer. However, other exciting applications in other fields are also not ruled out for the use of nanobodies.
REFERENCES
1. ALLELE-Biotechnology. Nanoantibodies [Internet]. Nanobodies. 2020. Disponible en: https://www.allelebiotech.com/nanoantibodies
2. Mir MA, Mehraj U, Sheikh BA, Hamdani SS. Nanobodies: The "Magic Bullets” in therapeutics, drug delivery and diagnostics. Hum Antibodies. 13 de febrero de 2020;28(1):29-51.
3. Alvez R. Obtención de Nanobodies específicos contra ROR-1 y AgB de Echinococcus Granulosus [Internet]. Universidad de la República I; 2018. Disponible en: https://www.colibri.udelar.edu.uy/jspui/bitstream/20.500.12008/19174/1/uy24-19055.pdf
4. Mitch L. Mini-antibodies discovered in sharks and camels could lead to drugs for cancer and other diseases [Internet]. Science. 2018. Disponible en: https://www.sciencemag.org/news/2018/05/mini-antibodies-discovered-sharks-and-camels-could-lead-drugs-cancer-and-other-diseases
5. Stanfield RL, Dooley H, Flajnik MF, Wilson IA. Crystal Structure of a Shark Single-Domain Antibody V Region in Complex with Lysozyme. Science. 17 de septiembre de 2004;305(5691):1770-3.
6. Matamoros, Alcivar E Ivanova,, González, Avilés M Selena,. Study review of camelid and shark antibodies for biomedical and biotechnological applications. Bionatura. 15 de noviembre de 2021;6(4):2331-40.
7. Flajnik M, Deschacht N, Muyldermans S. A Case Of Convergence: Why Did a Simple Alternative toCanonical Antibodies Arise in Sharks and Camels. PLOS Biol [Internet]. 2011;9. Disponible en: https://journals.plos.org/plosbiology/article/citation?id=10.1371/journal.pbio.1001120
8. Hersh L. Nanobodies®: The «hot» new research tool. [Internet]. Nanobodies®: The «hot» new research tool. 2021. Disponible en: https://cobre.med.uky.edu/cobre-nanobody#:~:text=The%20nanobodies%20to%20your%20protein,as%20its%20sequence%20as%20deliverables.&text=In%20many%20cases%20our%20core,mammalian%20cells%20and%20purifying%20it.
9. Scarrone M, González-Techera A, Alvez-Rosado R, Delfin-Riela T, Modernell Á, González-Sapienza G, et al. Development of anti-human IgM nanobodies as universal reagents for general immunodiagnostics. New Biotechnol. septiembre de 2021;64:9-16.
10. Xu J, Kim AR, Cheloha RW, Fischer FA, Li JSS, Feng Y, et al. Protein visualization and manipulation in Drosophila through the use of epitope tags recognized by nanobodies. eLife. 25 de enero de 2022;11:e74326.
11. Cui P, Ji R, Ding F, Qi D, Gao H, Meng H, et al. A complete mitochondrial genome sequence of the wild two-humped camel (Camelus bactrianus ferus): an evolutionary history of camelidae. BMC Genomics. diciembre de 2007;8(1):241.
12. Agnew D. Chapter 7 - Camelidae. 2018;22.
13. López P. EL PLEISTOCENO DE LA CUENCA DE CALAMA. 2018;1. Disponible en: https://www.researchgate.net/publication/323845017_Camelidae
14. Fundación Heifer Ecuador. CAMELIDOS SUDAMERICANOS [Internet]. Escuala Nacional de agro ecología; 2018. Disponible en: http://www.heifer-ecuador.org/wp-content/uploads/2018/03/22.-Camelidos-sudamericanos.pdf
15. Marín JC, Zapata B, González BA, Bonacic C, Wheeler JC, Casey C, et al. Sistemática, taxonomía y domesticación de alpacas y llamas: nueva evidencia cromosómica y molecular. Rev Chil Hist Nat [Internet]. junio de 2007 [citado 4 de agosto de 2022];80(2). Disponible en: http://www.scielo.cl/scielo.php?script=sci_arttext&pid=S0716-078X2007000200001&lng=en&nrm=iso&tlng=en
16. Durand MDC, Paytán A. La milenaria familia camelidae y el Nevado Huaytapallana. Universidad Continental, editor. Nat Soc [Internet]. 30 de junio de 2018 [citado 30 de mayo de 2022];01(01). Disponible en: http://journals.continental.edu.pe/index.php/naturalezaysociedad/article/view/425
17. FAO. 3.5. CAMÉLIDOS [Internet]. CAMÉLIDOS. 1989. Disponible en: https://www.fao.org/3/v8300s/v8300s18.htm
18. Avilés D, Montero M, Barros-Rodriguez. LOS CAMÉLIDOS SUDAMERICANOS: PRODUCTOS Y SUBPRODUCTOS USADOS EN LA REGIÓN ANDINA SOUTH AMERICAN CAMELIDS: PRODUCTS AND SUB-PRODUCTS USED IN THE ANDEAN REGION. Actas Iberoam En Conserv Anim. mayo de 2018;(11):30-8.
19. Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Bajyana E, et al. Naturally occurring antibodies devid of ligth chains. Nature. 1993;363.
20. Khodabakhsh F, Behdani M, Rami A, Kazemi-Lomedasht F. Single-Domain Antibodies or Nanobodies: A Class of Next-Generation Antibodies. Int Rev Immunol. 3 de diciembre de 2018;37(6):316-22.
21. Muyldermans S. Applications of Nanobodies. Annu Rev Anim Biosci. 16 de febrero de 2021;9(1):401-21.
22. Chames P, Rothbauer U. Special Issue: Nanobody. Antibodies. 6 de marzo de 2020;9:4.
23. Boulenouar H, .Amar Y, Bouchoutrouch N, Faouzi MEA, Cherrah Y, Sefrioui H. Research Article Nanobodies and their medical applications. Genet Mol Res [Internet]. 2020 [citado 27 de junio de 2022];19(1). Disponible en: http://www.funpecrp.com.br/gmr/articles/year2020/vol19-1/pdf/gmr18452_-_nanobodies-and-their-medical-applications.pdf
24. World Healt Organization. Handbook : good laboratory practice (GLP) / UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases [Internet]. 2001. Disponible en: https://apps.who.int/iris/bitstream/handle/10665/66894/TDR_PRD_GLP_01.2.pdf?sequence=1&isAllowed=y
25. Elsevier Connect. Fases de desarrollo de un nuevo fármaco [Internet]. Fases de desarrollo de un nuevo fármaco. 2020. Disponible en: https://www.elsevier.com/es-es/connect/medicina/edu-fases-de-desarrollo-de-un-nuevo-farmaco
26. Guerrero GAM. Las fases en el desarrollo de nuevos medicamentos. Medigraphic. 2009;5.
27. Morrison C. Nanobody approval gives domain antibodies a boost. Nat Rev Drug Discov. julio de 2019;18(7):485-7.
28. Steeland S, Vandenbroucke RE, Libert C. Nanobodies as therapeutics: big opportunities for small antibodies. Drug Discov Today. julio de 2016;21(7):1076-113.
29. Janeway C, Travers P, Walport M. Immunobiology: The Immune System in Health and Disease. 5th edition. [Internet]. Grand Science; 2001. Disponible en: https://www.ncbi.nlm.nih.gov/books/NBK27144/#:~:text=The%20IgG%20antibody%20molecule%20is,a%20flexible%20Y%2Dshaped%20structure
30. Gosnell W, Kramer K, Yamaga K. Immunology: Antibody Basics [Internet]. John A. Burns School of Medicine; 2008. Disponible en: https://jabsom.hawaii.edu/docs/Antibodies_Instructional_Module.pdf
31. Mitchell LS, Colwell LJ. Comparative analysis of nanobody sequence and structure data. Proteins Struct Funct Bioinforma. julio de 2018;86(7):697-706.
32. Schumacher D, Helma J, Schneider AFL, Leonhardt H, Hackenberger CPR. Nanobodies: Chemical Functionalization Strategies and Intracellular Applications. Angew Chem Int Ed. 23 de febrero de 2018;57(9):2314-33.
33. Kunz P, Zinner K, Mücke N, Bartoschik T, Muyldermans S, Hoheisel JD. The structural basis of nanobody unfolding reversibility and thermoresistance. Sci Rep. diciembre de 2018;8(1):7934.
34. Gauhar A, Privezentzev CV, Demydchuk M, Gerlza T, Rieger J, Kungl AJ, et al. Single domain shark VNAR antibodies neutralize SARS‐CoV‐2 infection in vitro. FASEB J [Internet]. noviembre de 2021 [citado 19 de agosto de 2022];35(11). Disponible en: https://onlinelibrary.wiley.com/doi/10.1096/fj.202100986RR
35. Cheong WS, Leow CY, Abdul Majeed AB, Leow CH. Diagnostic and therapeutic potential of shark variable new antigen receptor (VNAR) single domain antibody. Int J Biol Macromol. marzo de 2020;147:369-75.
36. Ryding S. VHH Antibodies (Nanobodies) Advantages and Limitations [Internet]. AZO Life Science. 2021. Disponible en: https://www.azolifesciences.com/article/VHH-Antibodies-(Nanobodies)-Advantages-and-Limitations.aspx
37. Ruano-Gallego D, Fraile S, Gutierrez C, Fernández LÁ. Screening and purification of nanobodies from E. coli culture supernatants using the hemolysin secretion system. Microb Cell Factories [Internet]. 11 de marzo de 2019 [citado 15 de mayo de 2021];18. Disponible en: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6410518/
38. Sandin S, Öfverstedt LG, Wikström AC, Wrange Ö, Skoglund U. Structure and Flexibility of Individual Immunoglobulin G Molecules in Solution. Structure. marzo de 2004;12(3):409-15.
39. Gettemans J, De Dobbelaer B. Transforming nanobodies into high-precision tools for protein function analysis. Am J Physiol-Cell Physiol. 1 de febrero de 2021;320(2):C195-215.
40. Tang Q, Owens RJ, Naismith JH. Structural Biology of Nanobodies against the Spike Protein of SARS-CoV-2. Viruses. 3 de noviembre de 2021;13(11):2214.
41. Cohen S. Antibody structure. J Clin Pathol. 1 de enero de 1975;s1-6(1):1-7.
42. Kunz P, Flock T, Soler N, Zaiss M, Vincke C, Sterckx Y, et al. Exploiting sequence and stability information for directing nanobody stability engineering. Biochim Biophys Acta BBA - Gen Subj. septiembre de 2017;1861(9):2196-205.
43. vinayagam M. Antibody Structure & Function [Internet]. Thiruvalluvar University; 2002. Disponible en: https://www.academia.edu/4978816/Antibody_Structure_and_Function
44. Kanmert D. Structure and interactions of human IgG-Fc. [Link??ping]: Department of Physics, Chemistry and Biology, Link??ping University; 2011.
45. Feige MJ, Hendershot LM, Buchner J. How antibodies fold. Trends Biochem Sci. abril de 2010;35(4):189-98.
46. Jovčevska I, Muyldermans S. The Therapeutic Potential of Nanobodies. Biodrugs. 2019;11-26.
47. Shen Z, Xiang Y, Vergara S, Chen A, Xiao Z, Santiago U, et al. A resource of high-quality and versatile nanobodies for drug delivery. iScience. septiembre de 2021;24(9):103014.
48. Shen Z, Xiang Y, Vegara S, Chen A, Xiao Z, Santiago U, et al. A robust and versatile nanobody platform for drug delivery [Internet]. Bioengineering; 2020 ago [citado 4 de agosto de 2022]. Disponible en: http://biorxiv.org/lookup/doi/10.1101/2020.08.19.257725
49. Schoonooghe S, Laoui D, Van Ginderachter JA, Devoogdt N, Lahoutte T, De Baetselier P, et al. Novel applications of nanobodies for in vivo bio-imaging of inflamed tissues in inflammatory diseases and cancer. Immunobiology. diciembre de 2012;217(12):1266-72.
50. Ackaert C, Smiejkowska N, Xavier C, Sterckx YGJ, Denies S, Stijlemans B, et al. Immunogenicity Risk Profile of Nanobodies. Front Immunol. 9 de marzo de 2021;12:632687.
51. Kang W, Ding C, Zheng D, Ma X, Yi L, Tong X, et al. Nanobody Conjugates for Targeted Cancer Therapy and Imaging. Technol Cancer Res Treat. 1 de enero de 2021;20:153303382110101.
52. Bannas P, Hambach J, Koch-Nolte F. Nanobodies and Nanobody-Based Human Heavy Chain Antibodies As Antitumor Therapeutics. Front Immunol. 22 de noviembre de 2017;8:1603.
53. Arbabi-Ghahroudi M. Camelid Single-Domain Antibodies: Historical Perspective and Future Outlook. Front Immunol. 20 de noviembre de 2017;8:1589.
54. Sathyajith D. What are Camelid Antibodies? News Med LifeScience. 23 de enero de 2022;3.
55. Fernandes CFC, Pereira S dos S, Luiz MB, Zuliani JP, Furtado GP, Stabeli RG. Camelid Single-Domain Antibodies As an Alternative to Overcome Challenges Related to the Prevention, Detection, and Control of Neglected Tropical Diseases. Front Immunol. 9 de junio de 2017;8:653.
56. Absolute Antibody. Antibody Fragments [Internet]. Antibody Enginnering. 2022. Disponible en: https://absoluteantibody.com/antibody-resources/antibody-engineering/antibody-fragments/
57. Bao G, Tang M, Zhao J, Zhu X. Nanobody: a promising toolkit for molecular imaging and disease therapy. EJNMMI Res. diciembre de 2021;11(1):6.
58. Asaadi Y, Jouneghani FF, Janani S, Rahbarizadeh F. A comprehensive comparison between camelid nanobodies and single chain variable fragments. Biomark Res. diciembre de 2021;9(1):87.
59. Payandeh Z, Kofeiti A, Sefid F. Nanobody structure analysis and determination of the functional conserve amino acid with bioinformatic tools. 2015;8.
60. Zielonka S, Empting M, Grzeschik J, Könning D, Barelle CJ, Kolmar H. Structural insights and biomedical potential of IgNAR scaffolds from sharks. mAbs. 2 de enero de 2015;7(1):15-25.
61. Protein Data Bank. 1MEL [Internet]. CRYSTAL STRUCTURE OF A CAMEL SINGLE-DOMAIN VH ANTIBODY FRAGMENT IN COMPLEX WITH LYSOZYME. 1997. Disponible en: https://www.rcsb.org/structure/1mel
62. Protein Data Bank. 1MLC [Internet]. MONOCLONAL ANTIBODY FAB D44.1 RAISED AGAINST CHICKEN EGG-WHITE LYSOZYME COMPLEXED WITH LYSOZYME. 1995. Disponible en: https://www.rcsb.org/structure/1mlc
63. Protein Data Bank. 1T6V [Internet]. Crystal structure analysis of the nurse shark new antigen receptor (NAR) variable domain in complex with lysozyme. 2004. Disponible en: https://www.rcsb.org/structure/1T6V
64. Muyldermans S. A guide to generation and design of nanobodies. FEBS J. 2021;288:2084-102.
65. Olichon A, de Marco A. Preparation of a Naïve Library of Camelid Single Domain Antibodies. En: Saerens D, Muyldermans S, editores. Single Domain Antibodies [Internet]. Totowa, NJ: Humana Press; 2012 [citado 4 de agosto de 2022]. p. 65-78. (Methods in Molecular Biology; vol. 911). Disponible en: http://link.springer.com/10.1007/978-1-61779-968-6_5
66. Yan J, Wang P, Zhu M, Li G, Romão E, Xiong S, et al. Characterization and applications of Nanobodies against human procalcitonin selected from a novel naïve Nanobody phage display library. J Nanobiotechnology. diciembre de 2015;13(1):33.
67. Yan J, Li G, Hu Y, Ou W, Wan Y. Construction of a synthetic phage-displayed Nanobody library with CDR3 regions randomized by trinucleotide cassettes for diagnostic applications. J Transl Med. diciembre de 2014;12(1):343.
68. Liu B, Yang D. Easily Established and Multifunctional Synthetic Nanobody Libraries as Research Tools. Int J Mol Sci. 27 de enero de 2022;23(3):1482.
69. Xiang Y, Sang Z, Bitton L, Xu J, Liu Y, Schneidman-Duhovny D, et al. Integrative proteomics identifies thousands of distinct, multi-epitope, and high-affinity nanobodies. Cell Syst. marzo de 2021;12(3):220-234.e9.
70. Zupancic JM, Desai AA, Tessier PM. Facile isolation of high-affinity nanobodies from synthetic libraries using CDR-swapping mutagenesis. STAR Protoc. marzo de 2022;3(1):101101.
71. de Marco A. Recombinant expression of nanobodies and nanobody-derived immunoreagents. Protein Expr Purif. agosto de 2020;172:105645.
72. Njeru FN, Kusolwa PM. Nanobodies: their potential for applications in biotechnology, diagnosis and antiviral properties in Africa; focus on application in agriculture. Biotechnol Biotechnol Equip. 1 de enero de 2021;35(1):1331-42.
73. Fang Z, Cao D, Qiu J. Development and production of nanobodies specifically against green fluorescence protein. Appl Microbiol Biotechnol. junio de 2020;104(11):4837-48.
74. Pardon E, Laeremans T, Triest S, Rasmussen SGF, Wohlkönig A, Ruf A, et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc. marzo de 2014;9(3):674-93.
75. Pourasadi S, Gargari SLM, Rajabibazl M, Nazarian S. Efficient production of nanobodies against urease activity of Helicobacter pylori in Pichia pastoris. Turk J Med Sci. 2017;7.
76. Clackson T, Hoogenboom H, Griffiths A, Winter G. Making antibody fragments using phage display libraries. Lett Nat. 1991;624-8.
77. Deffar K, Shi H, Li L, Wang X, Zhu X. Nanobodies - the new concept in antibody engineering. 2009;8.
78. Yu J, Guo Y, Gu Y, Fan X, Li F, Song H, et al. A novel silk fibroin protein–based fusion system for enhancing the expression of nanobodies in Escherichia coli. Appl Microbiol Biotechnol. marzo de 2022;106(5-6):1967-77.
79. Kariuki CK, Magez S. Improving the yield of recalcitrant Nanobodies® by simple modifications to the standard protocol. Protein Expr Purif. septiembre de 2021;185:105906.
80. Li D, Huang H. Heterologous Expression of Nanobodies:a Recent Progress. China Biotechnol [Internet]. 25 de agosto de 2017; Disponible en: https://manu60.magtech.com.cn/biotech/EN/10.13523/j.cb.20170813
81. Iwaki T, Hara K, Umemura K. Nanobody production can be simplified by direct secretion from Escherichia coli. Protein Expr Purif. junio de 2020;170:105607.
82. Chen Q, Zhou Y, Yu J, Liu W, Li F, Xian M, et al. An efficient constitutive expression system for Anti-CEACAM5 nanobody production in the yeast Pichia pastoris. Protein Expr Purif. marzo de 2019;155:43-7.
83. Sheikholeslami F, Rasaee MJ, Shokrgozar MA, Dizaji MM, Rahbarizadeh F, Ahmadvande D. Isolation of a Novel Nanobody Against HER-2/ neu Using Phage Displays Technology. Lab Med. febrero de 2010;41(2):69-76.
84. Shokrollahi N, Habibi Anbouhi M, Jahanian-Najafabadi A, AliRahimi E, Behdani M. Expressing of Recombinant VEGFR2-specific Nanobody in Baculovirus Expression System. Iran J Biotechnol [Internet]. marzo de 2021 [citado 19 de agosto de 2022];19(1). Disponible en: https://doi.org/10.30498/IJB.2021.2783
85. Wang W, Yuan J, Jiang C. Applications of nanobodies in plant science and biotechnology. Plant Mol Biol. 10 de octubre de 2020;1-11.
86. Ortega-Monge C, Arce-Rodríguez N, Santamaría-Muñoz M, Chavarría-Rojas M, Rojas Salas MF, Baltodano Viales E, et al. Aplicaciones de los nanoanticuerpos en la medicina. Ars Pharm Internet. 21 de marzo de 2022;63(2):189-203.
87. Van Audenhove I, Gettemans J. Nanobodies as Versatile Tools to Understand, Diagnose, Visualize and Treat Cancer. EBioMedicine. junio de 2016;8:40-8.
88. Hosseindokht M, Bakherad H, Zare H. Nanobodies: a tool to open new horizons in diagnosis and treatment of prostate cancer. Cancer Cell Int. diciembre de 2021;21(1):580.
89. Sanaei M, Setayesh N, Sepehrizadeh Z, Mahdavi M, Yazdi MH. Nanobodies in Human Infections: Prevention, Detection, and Treatment. Immunol Invest. 16 de noviembre de 2020;49(8):875-96.
90. Hassanzadeh-Ghassabeh G, Devoogdt N, De Pauw P, Vincke C, Muyldermans S. Nanobodies and their potential applications. Nanomed. junio de 2013;8(6):1013-26.
91. De Vlaminck K, Romão E, Puttemans J, Pombo Antunes AR, Kancheva D, Scheyltjens I, et al. Imaging of Glioblastoma Tumor-Associated Myeloid Cells Using Nanobodies Targeting Signal Regulatory Protein Alpha. Front Immunol. 30 de noviembre de 2021;12:777524.
92. Gu K, Song Z, Zhou C, Ma P, Li C, Lu Q, et al. Development of nanobody-horseradish peroxidase-based sandwich ELISA to detect Salmonella Enteritidis in milk and in vivo colonization in chicken. J Nanobiotechnology. diciembre de 2022;20(1):167.
93. Girt GC, Lakshminarayanan A, Huo J, Dormon J, Norman C, Afrough B, et al. The use of nanobodies in a sensitive ELISA test for SARS-CoV-2 Spike 1 protein. R Soc Open Sci. septiembre de 2021;8(9):211016.
94. Keown JR, Zhu Z, Carrique L, Fan H, Walker AP, Serna Martin I, et al. Mapping inhibitory sites on the RNA polymerase of the 1918 pandemic influenza virus using nanobodies. Nat Commun. diciembre de 2022;13(1):251.
95. DeMarco S. Snakebite antivenoms step into the future. 10 de septiembre de 2022;14.
96. Garaicoechea L, Aguilar A, Parra GI, Bok M, Sosnovtsev SV, Canziani G, et al. Llama Nanoantibodies with Therapeutic Potential against Human Norovirus Diarrhea. Sestak K, editor. PLOS ONE. 12 de agosto de 2015;10(8):e0133665.
97. Laustsen A, Gutiérrez J, Knudsen C, Johansen K, Bermúdez-Méndez E, Cerni F, et al. Pros and cons of different therapeutic antibody formats for recombinant antivenom development. Toxicon [Internet]. 2018; Disponible en: https://www.sciencedirect.com/science/article/pii/S0041010118301144#bib101
98. Bailon H, Yaniro V, Cáceres O, Colque E, Leiva W, Padilla C, et al. Development of Nanobodies Against Hemorrhagic and Myotoxic Components of Bothrops atrox Snake Venom. Front Immunol. 7 de mayo de 2020;11:1-12.
99. Hmila I, Saerens D, Abderrazek RB, Vincke C, Abidi N, Benlasfar Z, et al. A bispecific nanobody to provide full protection against lethal scorpion envenoming. FASEB J. septiembre de 2010;24(9):3479-89.
100. Yavuz SŞ, Çeli̇Kyurt İK. An update of anti-viral treatment of COVID-19. Turk J Med Sci. 2021;19.
101. Aria H, Mahmoodi F, Ghaheh HS, Faranak mavandadnejad, Zare H, Heiat M, et al. Outlook of therapeutic and diagnostic competency of nanobodies against SARS-CoV-2: A systematic review. Anal Biochem. marzo de 2022;640:114546.
102. NIH Instituto Nacional del Cancer. ¿Qué es el cáncer? [Internet]. 2021. Disponible en: https://www.cancer.gov/espanol/cancer/naturaleza/que-es
103. Lippman SM, Hawk ET. Cancer Prevention: From 1727 to Milestones of the Past 100 Years. Cancer Res. 1 de julio de 2009;69(13):5269-84.
104. NIH National Cancer INstitute. Cancer Treatment [Internet]. 2021. Disponible en: https://www.cancer.gov/about-cancer/treatment#:~:text=Some%20people%20with%20cancer%20will,targeted%20therapy%2C%20or%20hormone%20therapy
105. Yang EY, Shah K. Nanobodies: Next Generation of Cancer Diagnostics and Therapeutics. Front Oncol. 23 de julio de 2020;10:1182.
106. Verhaar ER, Woodham AW, Ploegh HL. Nanobodies in cancer. Semin Immunol. febrero de 2021;52:101425.
107. Wang J, Kang G, Yuan H, Cao X, Huang H, de Marco A. Research Progress and Applications of Multivalent, Multispecific and Modified Nanobodies for Disease Treatment. Front Immunol. 18 de enero de 2022;12:838082.
108. World Healt Organization. Coronavirus [Internet]. 2020. Disponible en: https://www.who.int/es/health-topics/coronavirus#tab=tab_1
109. World Healt Organization. WHO Director-General’s opening remarks at the media briefing on COVID-19 - March 11 2020 [Internet]. 2021. Disponible en: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020
110. Rodriguez-Guerra M, Jadhav P, Vittorio TJ. Current treatment in COVID-19 disease: a rapid review. Drugs Context. 29 de enero de 2021;10:1-8.
111. Hanke L, Sheward DJ, Pankow A, Vidakovics LP, Karl V, Kim C, et al. Multivariate mining of an alpaca immune repertoire identifies potent cross-neutralizing SARS-CoV-2 nanobodies. Sci Adv. 25 de marzo de 2022;8(12):eabm0220.
112. Verkhivker G. Structural and Computational Studies of the SARS-CoV-2 Spike Protein Binding Mechanisms with Nanobodies: From Structure and Dynamics to Avidity-Driven Nanobody Engineering. Int J Mol Sci. 8 de marzo de 2022;23(6):2928.
113. Saied AA, Metwally AA, Alobo M, Shah J, Sharun K, Dhama K. Bovine-derived antibodies and camelid-derived nanobodies as biotherapeutic weapons against SARS-CoV-2 and its variants: A review article. Int J Surg. febrero de 2022;98:106233.
114. United Nations. Three possible evolutions for the coronavirus causing COVID-19 [Internet]. 2022. Disponible en: https://news.un.org/es/story/2022/03/1506482
115. Ceretta M, Ramos B. Desarrollo de una estrategia para la obtención de nanobodies contra las toxinas botulínicas A y B de Clostridium botulinum [Internet]. Universidad ORT Uruguay; 2018. Disponible en: https://dspace.ort.edu.uy/bitstream/handle/20.500.11968/3919/Material%20completo.pdf?sequence=-1&isAllowed=y
116. Abbas A, Lichtman A, Pillai S. Celular and Molecular Immunology. novena. Philadelphia: Elsevier; 2017.
117. NIH. Glossary of HIV/AIDS-Related Terms. 2021;202.
118. Allmen D von, Shochat S. PRINCIPLES OF ADJUVANT THERAPY IN CHILDHOOD CANCER. En: Ashcraft’s Pediatric Surgery [Internet]. Elsevier; 2010 [citado 4 de agosto de 2022]. p. 837-52. Disponible en: https://linkinghub.elsevier.com/retrieve/pii/B9781416061274000665
119. Adel M Z, Abdullah A AD, Markus S, Ahmed A A, Emad M S, Basem S A, et al. Cloning and characterisation of nanobodies against the coat protein of Zucchini yellow mosaic virus. Plant Prot Sci. 25 de agosto de 2018;54(No. 4):215-21.
120. Hemmer C, Djennane S, Ackerer L, Hleibieh K, Marmonier A, Gersch S, et al. Nanobody‐mediated resistance to Grapevine fanleaf virus in plants. Plant Biotechnol J [Internet]. 2017;16. Disponible en: https://onlinelibrary.wiley.com/doi/full/10.1111/pbi.12819
121. He J, Tian J, Xu J, Wang K, Li J, Gee SJ, et al. Strong and Oriented Conjugation of Nanobodies onto Magnetosomes for the Development of a Rapid Immunomagnetic Assay for the Environmental Detection of Tetrabromobisphenol-A. Anal Bioanal Chem. octubre de 2018;410(25):6633-42.
122. Peltomaa R, Benito-Peña E, Moreno-Bondi MC. Bioinspired recognition elements for mycotoxin sensors. Anal Bioanal Chem. enero de 2018;410(3):747-71.
123. Wang Y, Fan Z, Shao L, Kong X, Hou X, Tian D, et al. Nanobody-derived nanobiotechnology tool kits for diverse biomedical and biotechnology applications. Int J Nanomedicine. julio de 2016;Volume 11:3287-303.
124. Janke BH. Influenza A virus infections in swine: pathogenesis and diagnosis. Vet Pathol. marzo de 2014;51(2):410-26.
125. Pinto Torres JE, Goossens J, Ding J, Li Z, Lu S, Vertommen D, et al. Development of a Nanobody-based lateral flow assay to detect active Trypanosoma congolense infections. Sci Rep [Internet]. 13 de junio de 2018 [citado 22 de mayo de 2021];8. Disponible en: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5998082/
126. Rexroad C, Vallet J, Matukumalli LK, Reecy J, Bickhart D, Blackburn H, et al. Genome to Phenome: Improving Animal Health, Production, and Well-Being – A New USDA Blueprint for Animal Genome Research 2018–2027. Front Genet [Internet]. 2019 [citado 16 de mayo de 2021];10. Disponible en: https://www.frontiersin.org/articles/10.3389/fgene.2019.00327/full?report=reader
127. Deckers N, Saerens D, Kanobana K, Conrath K, Victor B, Wernery U, et al. Nanobodies, a promising tool for species-specific diagnosis of Taenia solium cysticercosis. Int J Parasitol. abril de 2009;39(5):625-33.
128. Virdi V, Coddens A, De Buck S, Millet S, Goddeeris BM, Cox E, et al. Orally fed seeds producing designer IgAs protect weaned piglets against enterotoxigenic Escherichia coli infection. Proc Natl Acad Sci U S A. 16 de julio de 2013;110(29):11809-14.
129. Tang KL, Caffrey NP, Nóbrega DB, Cork SC, Ronksley PE, Barkema HW, et al. Restricting the use of antibiotics in food-producing animals and its associations with antibiotic resistance in food-producing animals and human beings: a systematic review and meta-analysis. Lancet Planet Health. 1 de noviembre de 2017;1(8):e316-27.
130. Yang J, Si R, Wu G, Wang Y, Fang R, Liu F, et al. Preparation of Specific Nanobodies and Their Application in the Rapid Detection of Nodularin-R in Water Samples. Foods. 10 de noviembre de 2021;10(11):2758.
131. Otero J. Los anticuerpos y su papel como herramientas analíticas en los ensayos inmunoenzimáticos. Rev Cubana Med Trop. agosto de 2010;62(2):85-92.
132. Nykiel-Szymańska J, Stolarek P, Bernat P. Elimination and detoxification of 2,4-D by Umbelopsis isabellina with the involvement of cytochrome P450. Environ Sci Pollut Res Int. 2018;25(3):2738-43.
133. Huang L, Muyldermans S, Saerens D. Nanobodies ® : proficient tools in diagnostics. Expert Rev Mol Diagn. septiembre de 2010;10(6):777-85.
134. Steyaert J, Pardon E, Wohlkönig A, Zögg, Kalichuk V, De Keyser P, et al. NANOBODY EXCHANGE CHROMATOGRAPHY [Internet]. p. 146. Disponible en: https://patentimages.storage.googleapis.com/63/2c/94/15a6e50d9a4a9f/WO2021123360A1.pdf
135. McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol. marzo de 2018;25(3):289-96.
136. Uchanski T, Pardon E, Steyaert J. Nanobodies to study protein conformational states. Curr Opin Struct Biol. 2020;60.
137. Beghein E, Gettemans J. Nanobody Technology: A Versatile Toolkit for Microscopic Imaging, Protein–Protein Interaction Analysis, and Protein Function Exploration. Front Immunol. 4 de julio de 2017;8:771.
138. Marschall ALJ, Dübel S. Antibodies inside of a cell can change its outside: Can intrabodies provide a new therapeutic paradigm? Comput Struct Biotechnol J. 2016;14:304-8.
139. Dong JX, Lee Y, Kirmiz M, Palacio S, Dumitras C, Moreno CM, et al. A toolbox of nanobodies developed and validated for use as intrabodies and nanoscale immunolabels in mammalian brain neurons. eLife. 30 de septiembre de 2019;8:e48750.
140. Trimmer JS. Genetically encoded intrabodies as high-precision tools to visualize and manipulate neuronal function. Semin Cell Dev Biol. junio de 2022;126:117-24.
141. Lo ASY, Zhu Q, Marasco WA. Intracellular Antibodies (Intrabodies) and Their Therapeutic Potential. En: Chernajovsky Y, Nissim A, editores. Therapeutic Antibodies [Internet]. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008 [citado 4 de agosto de 2022]. p. 343-73. (Starke K. Handbook of Experimental Pharmacology; vol. 181). Disponible en: http://link.springer.com/10.1007/978-3-540-73259-4_15
142. Lecerf JM, Shirley TL, Zhu Q, Kazantsev A, Amersdorfer P, Housman DE, et al. Human single-chain Fv intrabodies counteract in situ huntingtin aggregation in cellular models of Huntington’s disease. Proc Natl Acad Sci. 10 de abril de 2001;98(8):4764-9.
143. Marzilli AM, McMahan JB, Ngo JT. Precision control of intrabodies in live cells. Nat Methods. marzo de 2020;17(3):259-60.
144. NIH. Coronavirus Disease 2019 (COVID-19) Treatment Guidelines [Internet]. 2022. Disponible en: https://www.ncbi.nlm.nih.gov/books/NBK570371/pdf/Bookshelf_NBK570371.pdf
145. Becker RC. Covid-19 treatment update: follow the scientific evidence. J Thromb Thrombolysis. julio de 2020;50(1):43-53.
146. Yu B, Li S, Tabata T, Wang N, Kumar R, Liu J, et al. Accelerating PERx Reaction Enables Covalent Nanobodies for Potent Neutralization of SARS-Cov-2 and Variants. BioRxiv Prepr [Internet]. 2022;1. Disponible en: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8936105/
147. Shi Z, Li X, Wang L, Sun Z, Zhang H, Chen X, et al. Structural basis of nanobodies neutralizing SARS-CoV-2 variants. Structure. mayo de 2022;30(5):707-720.e5.
148. Wagner T, Schnepf D, Beer J, Ruetalo N, Klingel K, Kaiser P, et al. Biparatopic nanobodies protect mice from lethalchallenge with SARS-CoV-2variants of concern. EMBO Rep. 20 de diciembre de 2021;4(23):12.
149. Torres O. REPTILES DEL ECUADOR [Internet]. Bioweb. 2021. Disponible en: https://bioweb.bio/faunaweb/reptiliaweb/
150. Ministerio de Salud Pública del Ecuador. Manejo clínico del envenenamiento por mordeduras de serpientes venenosas y picaduras de escorpiones [Internet]. Puyo: Gobierno de la república del ecuador; 2017. Disponible en: http://hgp.gob.ec/index.html/images/Protocolo%20serpientes%202017.pdf
151. Fan HW, Natal Vigilato MA, Augusto Pompei JC, Gutiérrez JM, en representación de la Red de Laboratorios Públicos Productores de Antivenenos de América Latina (RELAPA).(Los autores de la RELAPA y sus afiliaciones se mencionan al final del manuscrito.). Situación de los laboratorios públicos productores de antivenenos en América Latina. Rev Panam Salud Pública. 19 de noviembre de 2019;43:1.
152. Ministerio del Ambiente Ecuador. NORMA DE CALIDAD AMBIENTAL Y DE DESCARGA DE EFLUENTES : RECURSO AGUA. 2015.
153. Jaramillo-Feijoo L, Real-Cotto J, Tanca-Campozano J, Puga-Peña G, Quinto-Briones R. Incidencia y mortalidad del cáncer, en Hospital Solca - Guayaquil. :6.
154. National Tumor Registry (RNT). Cancer epidemiology in Quito 2011-2015 [Internet]. 2019. Disponible en: https://issuu.com/solcaquito/docs/epidemiolog_a_del_c_ncer_en_quito_2011-2015
Citation: Ortega C P, Rivera L M, Trujillo L E. Production of nanobodies in Andean camelids and their most common applications: A general review in the medical field. Revis Bionatura 2023;8 (2) 13. http://dx.doi.org/10.21931/RB/2023.08.02.13
