2022.07.03.27
Files > Volume 7 > Vol 7 No 3 2022
Analysis a number of Quantitative Traits and Genetic Variation of Different Generation of Wheat (Tritecum aestivum) by using RAPD-PCR
Raed Salem Alsaffar
Department of Biology, Faculty of Science, University of Mosul
Corresponding author. [email protected]
Available from: http://dx.doi.org/10.21931/RB/2022.07.03.27
ABSTRACT
RAPD-PCR genetic markers were used to assess genetic variation in wheat plants and connections among six wheat genotypes. Four random primers produced 140 DNA fragments, averaging 6.7 identifiable bands per primer. Among the six genotypes, 85 pieces (44.64 percent) were polymorphic. Several RAPD marker bands had distinct signify recurrence patterns that thing differently amongst germplasm of wheat plants groupings. Within-community genetic variation accounted for 78 to 89 percent of the overall variance. Wheat genotypes may be characterized and classified using RAPD analysis. These findings will be a benefit in wheat-producing offspring efforts in the future.
Keywords. Barley, Genetic variation, RAPD-PCR.
INTRODUCTION
Wheat grains are the world's most critical cultivated plants and humanity's most crucial nutritional staple1. Wheat breeding changed forever with the development of short-statured, fertilizer-responsive wheat cultivars. As a result, grain production potential has increased significantly2,3,4.
Wheat is cultivated on 40% of Pakistan's cultivable land, with an average output of 2495 kg/ha5,6. In comparison to other agricultural countries, this is relatively low. Understanding local variations' genetics and genomic structure using molecular markers are beneficial for breeding purposes7,8. The first stage in wheat improvement, like any other crop species, is a thorough examination of the local resources, including the collection, appraisal, and molecular characterization of germplasm lines. Crop development efforts might benefit significantly from understanding germplasm diversity and genetic linkages among breeding materials. Data on germplasm variety and genetic relatedness among high-quality breeding stock are crucial in plant breeding6,5,4,3.
Although a varied genetic foundation has recently been recommended for wheat disease resistance4, Future breeding efforts will also rely on the availability of genetic diversity to enhance productivity5.
As a result, to attain self-sufficiency and sustainability, cultivars with a broad genetic foundation must be developed6.
Breeders can better grasp the evolutionary links between accessions if they know diversity trends7.
Traditionally, variations in morphological and agronomic characteristics have been used to analyze wheat genetic diversity and pedigree information8.
The focus today is on collecting genetic diversity within and between accessions. SSR, RFLP, RAPD and AFLP analyses are the finest approaches for achieving this9.
The current method for assessing genetic diversity within germplasm collections is RAPD for cultivar identification utilizing DNA profiling10.
PCR depends on the RAPD technique as genetic markers are the prevailing marker widely utilized in genetic mapping11. And the identification of loci associated with various phenotypes12.
RAPD – PCR technique has been utilized for genetic variation studies in various plants because of its technical simplicity and rapidity13.
This study's goal was to compare the qualities of wheat plant phylogenetic groups depending on seed traits clustering to phylogenetic groupings based on RAPD-PCR clustered using UPGMA14.
MATERIAL AND METHODS
DNA extraction from the seed of Wheat
DNA of seed was isolated from 6 different wheat generation (Triticum aestivum L.) immature leaves. At 25 °C in the dark, all generations were planted in plastic pots (200 ml). The generated kit was used to extract DNA from 15-day-old seedlings11. Then the DNA isolated was kept at -80 C. To utilize for PCR amplifications.

Table 1. The 4 RAPD primers used are shown and PCR conditions
RESULT AND DISCUSSION
Following electrophoresis in SB buffer, the RAPD-PCR bands were specified on 2 percent (w/v) agarose gel electrophoresis and observed under UV light after Red Safe staining.
All amplifications were done twice to ensure that the amplification of scored fragments was repeatable. All visible RAPD fragments were enumerated for each primer, and strong polymorphic bands were graded as present (1) or absent (0). The number of polymorphic bands for each primer was determined.

Figure 1. The RAPD-PCR product for 6 generations of Wheat with (GLE-01) Primer and M ladder 100 bp

Figure 2. The RAPD-PCR product for 6 generations of Wheat with (GLE-02) Primer and M ladder 100 bp
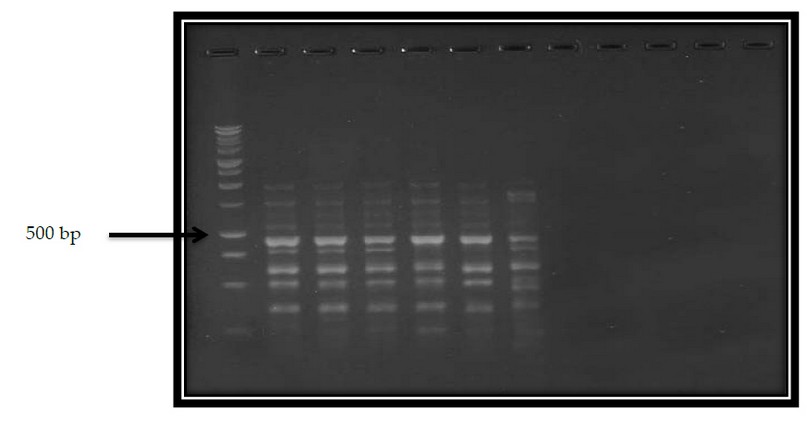
Figure 3. The RAPD-PCR product for 6 generations of Wheat with (GLE-03) Primer and M ladder 100 bp

Figure 4. The RAPD-PCR product for 6 generations of Wheat with (GLE-04) Primer and M ladder 100 bp
A dendrogram built with UPGMA clustering was used to assess genetic connections between generations.

Figure 5. The Dendrogram of 6 different wheat generations developed from RAPD-PCR data utilizes unweighted pair grouping of arithmetic means (UPGMA).
The 4 primers produced 100 DNA fragments, with an average of 6.7 bands for each primer. 45.22 percent of the amplified fragments were polymorphic. All the primers produced 4 to 11 amplification products ranging in size from 0.27 to 3.6 kb. The primer sequence solely determined the size and quantity of DNA fragments (fir15(.With various primers, the degree of polymorphism varied between generations. These findings show that RAPD-PCR markers offered helpful information for wheat generation identification. Momal-2002 produced the most DNA amplified bands (85) of the 10 genotypes examined, whereas line CIM-31 produced the fewest16.
Several variables can affect the repeatability of the RAPD method, including primer sequence, template quality and amount, thermocycler type, and Taq-polymerase activity11,14.
The implementation of a defined RAPD procedure, on the other hand, can assure that RAPD patterns are repeatable. In each repeat, all of the amplified bands were identical. A similarity matrix was generated using multivariate analysis to assess genetic diversity among wheat generations18.
UPGMA analysis was used to create a dendrogram (Fig. 5) to identify the grouping of the wheat generation using these similarity coefficients.
Just 8670-3 was found in a second cluster, the most different generation examined, with only 79.2 percent similarity to the other genotypes in our research9,10.
RAPD analysis proved highly successful in detecting genetic diversity among wheat generations, despite the small genetic basis among the 6 generations employed here, and can be used to create DNA fingerprints for identifying different varieties13.
The genetic isolation between generations (Fig. 5) might be due to a few seed firms' closed guild marketing procedures.
CONCLUSION
The ramifications of these discoveries for wheat plant breeding are significant. These genotypes' genetic material links might aid in selecting genetically diverse parents for germplasm production. The discovery of a minor genetic alteration in Wheat emphasizes the need to expand the genetic base of wheat breeding materials. This genetic diversity index might help researchers decide which parents to employ for genome mapping 8,12.
Funding: self-funding
Acknowledgments: In this section, we acknowledge any person who supports us in completing this project.
Conflicts of Interest: there is no conflict
REFERENCE
1. Elena Benavente 1 , Francisco Manzano-Agugliaro 2 and Estela Gimenez
2. Khatri, N., et al., Effect of Different Wheat Variety and Sowing Methods on Grain Yield of Wheat under Bhairahawa Condition of Nepal. International Journal of Life Sciences and Biotechnology, 2019. 2(3): p. 175-182.
3. Ozyigit, I.I., et al., The effects of cadmium on growth, some anatomical and physiological parameters of Wheat (Triticum aestivum L.). International Journal of Life Sciences and Biotechnology, 2021. 4(2): p. 235- 253. DOI: 10.38001/ijlsb.833553
4. Bai, G., Guo, P., Kolb, F.L.,. Genetic relationships among head blight resistant cultivars of Wheat assessed on the basis of molecular markers. Crop Science, 2003, 43, 498– 507.
5. Chalmers, J.K., Campbell, A.W., Krestschmer, J., Karakousis, A., Henschke, P.H., Pierens, S., Harker, N., Pallotta, M., Cornish, G.B., Shariflou, M.R., Rampling, L.R., McLauchlan, A., Daggard, G., Sharp, P.J., Holton, T.A., Sutherland, M.W., Appels, R., Langridge, P.Construction of three linkage maps in bread wheat, Triticum aestivium L. Australian Journal of Agriculture Research , 2001,52, 1089–1119.
6. Hernendez, P., Rosa, R., Rallo, L., Dorado, G., Martin, A. Development of SCAR markers in Olive (Olea europaea) by direct sequencing of RAPD products: application in olive germplasm evaluation and mapping. Theoretical and Applied Genetics , 2001,103, 788– 791.
7. Nabulsi, I., Al-Safaid,, Ali, N., Arabi, MIE Evaluation of some garlic (Allium sativum L.) mutants resistant to white rot disease by RAPD analysis. Annals of Applied Biology , 2001,138, 197–202.
8. Sun, G., Bond, M., Nass, H., Martin, R., Dong, Z. DNA polymorphisms in spring wheat cultivars and lines with different level of Fusarium resistance. Theoretical and Applied Genetics , 2003,106, 1059– 1067.
9. Marakli, S., A Brief Review of Molecular Markers to Analyse Medicinally Important Plants. International Journal of Life Sciences and Biotechnology, 2018. 1(1): p. 29-36.
10. Zhu, Y., Chen, H., Fan, J., Wanf, Y., Li, Y., Chen, J., Fan, J., Yang, S., Hu, L., Leung, H., Mew, T.W., Teng, P.S., Wang, Z., Mundt, C.C. Genetic diversity and disease control in rice. Nature , 2000,406, 718– 722.
11. Bhutta M, Akhtar J, Ibrahim M. and Shahzad A. Genetic variation between Pakistani Wheat (Triticum aestivum L.) genotypes as revealed by Random Amplified Polymorphic DNA (RAPD) markers. South African Journal of Botany , 2006,72 (2006) 280 – 283.
12. Bernard, R.L., Cremeens, C.R., Cooper, R.L., Collins, F.L., Krober, O.A., Athow, K.L., Laviolette, F.A., Cobe, C.J., Belson, R.L. Evaluation of the USDA Soybean germplasm collection: maturity groups 000 to IV (FC01.547–PI266.807). Technical Bulletin-United States Department of Agriculture, 1998,1844.
13. Demek, T., Lynch, D.R., Kawchuk, L.M., Kozub, G.C., Armstrong, J.D. Genetic diversity of potato determined by random amplified polymorphic DNA analysis. Plant Cell Reports, 1996, 15, 662– 667.
14. Paull, J.G., Chalmers, K.J., Karakousis, A., Kretschmer, J.M., Manning, S., Langridge, P. Genetic diversity in Australian wheat varieties and breeding material based on RFLP data. Theoretical and Applied Genetics, 1998, 96, 435– 446.
15. Tahir, N.A. Assesment of genetic diversity among wheat varities in sulaimanyah using random amplified polymorphic DNA (RAPD) analysis. Jordan Journal of Biological Sciences, 2008, 1(4): 159-164
16. Bhutta, W. M. Biochemical and molecular characterization of wheat genotypes determined by RAPD Analysis. Acta Agriculturae Scandinavica, Section B- Plant Soil Sience, 2006, 57(4):335-341
17. Chen, H. B., J. M. Martin, M. Lavin, L. E. Talbert. Genetic diversity in hard red spring wheat based on sequence-tagged-site PCR markers. Crop Science, 1994,34(6): 1628-1632.
18. Bedo, Z., L. Szunics, L. Lang, Lu. Szunics, O. Veisz, I. Karsai, GY. Vida, P. Szücs, A. Juhasz, M. Gal, SZ. Bencze, M. Megyeri, K. Puskas, C. S. Horvath. Genetic diversity in durum wheat. Annual Wheat Newsletter, 2000, Vol:46. http://wheat.pw.usda.gov/gpages/awn/
19. Cao, W., P. Hucl, G. Scoles, R. N. Chibbar, P. N. Fox ,B. Skovmand. Cultivar identification and pedigree assessment of common Wheat based on RAPD analysis, 2002,Wheat Information Service Number 95: 29-35.
20. Abbas, S.J., SRU. Shah, G.Rasool ,A. Iqbal.Analysis of genetic diversity in Pakistani wheat varieties by using randomly amplified polymorphic DNA (RAPD) primers American-Eurasian Journal of Sustainable Agriculture, 2008 , 2(1): 29-33.
21. Altıntas¸ S., F. Toklu, S. Kafkas, B. Kilian, A. Brandolini, H. Özkan. Estimating genetic diversity in durum and 95 bread wheat cultivars from Turkey using AFLP and SAMPL markers. Plant Breeding , 2008,127: 9-14.
22. Motawei, M.I., A.A. Al-doss, K.A. Moustafa. Genetic diversity among selected wheat lines differing in heat tolerance using molecular markers. Journal of Food, Agriculture and Environment, 2007,5(1):180-183.
.
Received: 25 January 2022 / Accepted: 14 June 2022 / Published:15 Agoust 2022
Citation: R Salem Alsaffar. . Analysis a number of Quantitative Traits and Genetic Variation of Different Generation of Wheat (Tritecum aestivum) by using RAPD-PCR. Revis Bionatura 2022;7(3) 27. http://dx.doi.org/10.21931/RB/2022.07.03.27
