2022.07.01.10
Files > Volume 7 > Vol 7 No 1 2022
Assessment of genetic fidelity of lacy tree philodendron (Philodendron bipinnatifidum Schott ex Endl.) micro propagated plants
1Department of Botany and Microbiology, College of Science, King Saud University, Riyadh 11495, Saudi Arabia; [email protected] (MSA); [email protected] (AAA); [email protected] (DAS.)
2Plant Production Department, College of Food and Agriculture Sciences, King Saud University, Riyadh 11451, Saudi Arabia.
3Department of Horticulture, Faculty of Agriculture, Kafrelsheikh University, Kafr El-Sheikh 33516, Egypt.
4Ornamental and Floriculture Department, Horticulture Research Institute, Antoniades, Alexandria 21599, Egypt; [email protected]
* Correspondence: [email protected] (YHD.)
Available from: http://dx.doi.org/10.21931/RB/2022.07.01.10
ABSTRACT
Tissue culture is a potent means for producing clonally propagated plants. However, genetically identical regenerants are of great concern due to their economic consequences. Therefore, polymerase chain reaction (PCR)-based molecular markers are employed to detect somaclonal variations. In this study, the genetic fidelity of in vitro raised Philodendron bipinnatifidum clones, and their mother plant was tested using 11 randomly amplified polymorphic DNA (RAPD) markers. The RAPD decamers produced 92 amplicons with 8.4 bands ranging from 260-5000 bp. The bands varied from one to fifteen for primer 8 and primer 7, respectively. The genetic similarity between the micro propagated plantlets and mother plant of P. bipinnatifidum was nearly 100%, assuring uniformity and true-to-type regenerated plantlets for this commercially crucial ornamental plant.
Keywords. Araceae; genetic fidelity; micropropagation; polymorphism; RAPD analysis.
INTRODUCTION
Rapid clonal propagation is one of the most extensively used approaches of biotechnology. In vitro plants undergo several subculture cycles before their rooting, and acclimatization and cytogenetic variations may occur in this long process1. Somaclonal variations can be a valuable source for new genetic material2 and might be advantageous for crop improvement 3. However, its occurrence in micro propagated plants results in undesirable plant off-types, limiting its applications in the tissue culture industry. Therefore, the most critical concern has been the maintenance of genetic uniformity and the production of true-to-type plants. The influence of in vitro culture conditions i.e., culture media, type of explant, and subculture cycles, may lead to genomic variation in the regenerated plantlets. Therefore, assessment of genetic stability is a prerequisite while performing micropropagation 4.
Several methods were available to assess regenerated plants' genetic fidelity, such as morphological characteristics, cytological and flow cytometry analysis 5. DNA based molecular markers emerged as the most suitable genetic tools to determine the genetic variation among regenerated plants 6. Molecular markers such as randomly amplified polymorphic DNA (RAPD) 7, amplified fragment length polymorphism (AFLP) 8, simple sequence repeat (SSR) 9 and inter simple sequence repeats (ISSR)10,11are used for determining the genetic fidelity of micro propagated plants. Cost-effectiveness is a major checkpoint in micropropagation. Because of the simplicity and cost effectiveness of RAPD technique, it is used for genetic stability analysis, amplifying the different regions of the genome 12 in several plant species 4, 6, 13, 14.
Lacy tree philodendron (Philodendron bipinnatifidum Schott Ex Endl.), formerly known as P. selloum K. Koch, is a self-heading ornamental plant species that can reach heights of 4–5 m and possesses deeply cut, green to dark green leaves that can grow up to 1 m in length. Conventional propagation does not encourage the multiplication of the species 15. Therefore, in vitro propagation methods can be utilized to facilitate the continuous generation of high-quantity and high-quality plant materials compared with traditional vegetative propagation. Our previous report established an efficient micropropagation protocol for lacy tree philodendron through axillary shoot regeneration with a 100% rooting and acclimatization 15. Therefore, the objective of this study was to assess the genetic fidelity of the regenerants to assure the production of genetically identical plantlets.
MATERIALS AND METHODS
Plant material
The mother plant of P. bipinnatifidum (Figure 1 A) and ten micro propagated plantlets through axillary shoot proliferation15 were randomly selected and used as plant material in this study. The axillary shoots were multiplied on MS medium 16 that contained 6-benzylaminopurine (1 mg·L-1) and indole-3-butyric acid (0.5 mg·L-1) for 6 wks (Figure 1 B). These in vitro shoots were kept in our laboratory for nine subculture cycles, in vitro plantlets rooted on MS medium that contained naphthalene acetic acid (1 mg·L-1) for 6 wks (Figure 1 C) and acclimatized in a 1:1 (v:v) mixture of peat moss and perlite for 30 d (Figure 1 D) as described by Alawaadh et al., 15. Young fresh leaves of the mother plant and clonal regenerants were thoroughly cleaned, washed with sterile distilled water, and kept in liquid nitrogen until molecular analysis.

Figure 1. Micropropagation of Philodendron bipinnatifidum (A) Mother plant, (B) Shoot multiplication in gelled culture, full-strength Murashige and Skoog medium that contained 6-benzylaminopurine (1 mg·L-1) and indole-3-butyric acid (0.5 mg·L-1), after 6 wks of culture, (C) In vitro plantlets rooted on Murashige and Skoog medium that contained naphthalene acetic acid (1 mg·L-1), after 6 wks of culture, (D) Acclimatized plantlet in a 1:1 (v:v) mixture of peat moss and perlite, after 30 d. (Unpublished photographs).
Genomic DNA isolation
Genomic DNA was isolated from fresh leaves of both mother plants and micro propagated plants by using the cetyltrimethylammonium bromide (CTAB) method 17. Approximately 100-150 mg of leaves were ground using 600 μL of preheated (65°C) extraction buffer (2% CTAB, 20 mM EDTA, 100 mM Tris-HCl, 1.4 M NaCl, 2% polyvinylpyrrolidone (PVP), 0.2% mercaptoethanol), transferred to a centrifuge tube (2 mL) and incubated for 30 min in a 65°C water bath, the samples were inverted every 5 min. 600 μL of chloroform-isoamyl alcohol (24: 1) was added and mixed by inverting the tubes carefully 10 times, and the cells were centrifuged at 12000 rpm for 10 min at room temperature. The supernatant was collected and carefully mixed with a two-thirds volume of ice-cold Isopropanol, and the DNA samples were collected by centrifuging for 10 min. RNaseA (10 μg/ml) was added to the 50 μl of TE buffer (10 mM Tris and 0.1 mM EDTA) before dissolving the DNA to remove any RNA in the preparation, and the mixture was incubated at 37 °C for 30 min. 100 μL and 750 μL of 3 M sodium acetate and absolute ice-cold ethanol were added. The DNA was collected by high-speed centrifugation for 10 min, and then carefully washed with ice-cold absolute and 70% ethanol and centrifuged at 12000 rpm for 10 min. Finally, the samples were dried at room temperature and dissolved in 50-100 μL of TE buffer. The quality and concentration of DNA were determined by EMPLEN photometer P330.
PCR conditions
11 decamer RAPD primers (Macrogen Korea) were used for DNA amplification. PCR reaction was carried out in a volume of 20 µL. PCR was performed as follows: 94°C for 5 min; followed by 35 cycles of 94°C for 1 min, specific annealing temperature (Ta) according to the primer sequence for 30 sec and 72°C for 3 min and the final extension step at 72°C for 10 min. Amplification was carried out in MJ Mini Bio RAD, thermal cycler in 25 µl reaction volume containing the following reagents: 1.0 µL of dNTPs (10 mM), 1.0 µL of MgCl2 (25 mM), 5 µL of 10x buffer, 1.0 µL of primer (10 pmol), 1.0 µL of DNA (25 ng µL-1), 0.3 µL of taq polymerase (5 u µL-1 ) and 15.7 dd.H2O.
Gel electrophoresis and genetic analysis
The RAPD products were electrophoresed in 1.5 % agarose gel containing Red safe dye in TAE buffer (40 mM Tris-acetate, 20 mM glacial acetic acid, 1 mM EDTA, pH 7) at 75 V. The gels were documented using a gel documentation system (UVITEC, UK) and according to an analysis by Phoretix program 1D gel analysis software version 4.01). Two repeats were performed to confirm the results. The bands obtained by scoring the RAPD profiles were treated as binary characters and coded accordingly (presence = 1, absence = 0). The genotypes showing similarity in their RAPD characteristics were grouped using UPGMA (Unweighted Paired Group with Arithmetic Average) The SPSS-10 package was used for statistical analysis.
RESULTS
Assessment of genetic fidelity was performed on the P. bipinnatifidum micro propagated plantlets using RAPD analysis with 11 RAPD primers (Table 1; Figure 2). The selected primers gave 92 scorable bands with an average of 5.8 bands ranging from 260-5000 bp. The bands varied from 1 to 15 for primer 8 and primer 7, respectively.
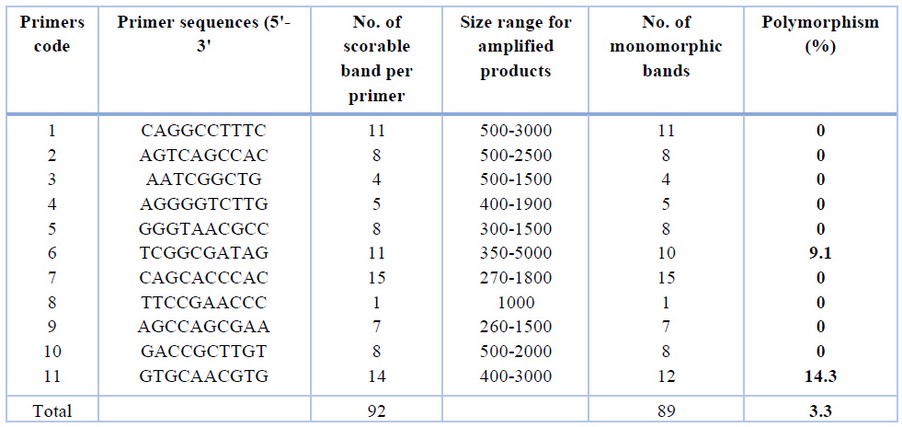
Table 1. RAPD primers were used for testing the genetic fidelity of Philodendron micro propagated plants.
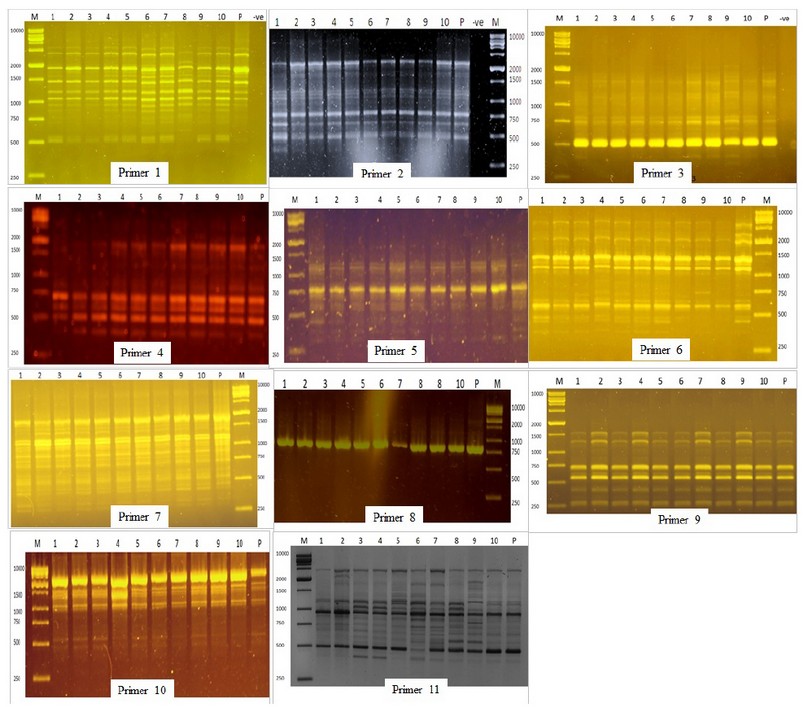
Figure 2. DNA Amplification pattern obtained with RAPD primers. Lane M- DNA ladder; Lane-P D.N.A. from mother plant; Lane 1-10 DNA from micropropagated plants.
RAPD amplification pattern obtained with all primers except for primers 6 and 11 revealed that all bands produced by micro propagated plants were monomorphic and similar to that of the mother plant. Primer 6 amplified 10 monomorphic bands and one unique polymorphic band (750 bp) produced by in vitro propagated clone 9. Primer 11 produced 14 scorable bands, out of which 12 were monomorphic while two were polymorphic. A unique band with a molecular size of 2400 bp was produced only in propagated clone 2, whereas micro propagated clones 3,4,5,6,7,8 produced a band with a molecular size of 1000 bp. Of the 11 primers analyzed, nine primers showed 0% polymorphism, while primers 6 and 11 showed 9.1% and 14.3% polymorphism, respectively. The genetic similarity between the micro propagated plantlets and the mother plants ranged from 0.98 to 1.00, with an average of 0.99 (Table 2).
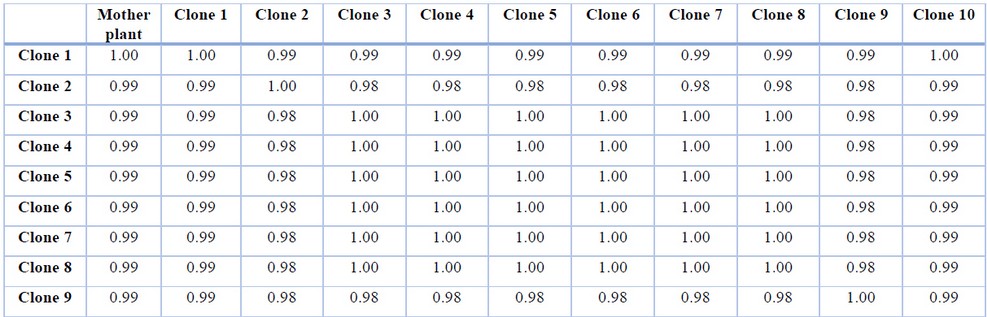
Table 2. Similarity coefficient among the screened plants based on RAPD analysis.
DISCUSSION
High genetic fidelity and true-to-type clones are critical for commercial micropropagation to maintain the essential characteristics of the mother plant 1. Phenotypic variations such as abnormal leaf shape and dwarf growth indicate off-type clones. However, the absence of these morphological abnormalities does not guarantee true-to-type regenerated plantlets; therefore, molecular markers should be utilized to assure their clonal fidelity. It has been reported that although in vitro regenerated Philodendron micans plantlets phenotypically resembled mother plants, they showed a high frequency of ploidy change suggests the occurrence of somaclonal variation 18. The author indicated that analysis of 20 randomly selected Philodendron micans plantlets using DNA flow cytometry showed that two were mixoploid. RAPD profiles of the randomly selected clones of Philodendron 'Xanadu' different ages compared to the mother plant were identical, thus assuring a genetic fidelity-maintained protocol for this commercially important plant 19. RAPD molecular marker was used to assess genetic fidelity in other micro propagated plant species belonging to the Araceae family, such as Aglaonema' Valentine’ 7 where all tested plantlets were highly uniform or identical to the mother plant. Conversely, for Anthurium Andreanum Bicolour ‘Agnihotri’, RAPD analysis revealed variations in band pattern of the regenerated plantlets 20.
Genetic variation is a common phenomenon observed in plants regenerated from tissue cultures 3. Various factors such as genotype, explant type, culture periods and growth regulators combinations and concentrations may disturb the internal polarity and physiology of the explants 21, 22 and could also influence the stability of tissue cultured plants 22, 23. Therefore, limiting the number of subculture cycles could maintain clonal characteristics. For example, DNA polymorphism (20%) was reported in protocorm like-bodies of Phalaenopsis gigantea after 20 wks of culture on a medium containing 4.5 μM thidiazuron and 65.5 μM chitosan while reducing the culture period to 16 wks resulted in no variations 24. However, the amenability to in vitro induced mutagenic effect is species-dependent. Borsai et al. 25 reported no polymorphism in micro propagated blackberry plants (Rubus fruticosus) for 12 subcultures during 30 months of in vitro multiplication.
CONCLUSIONS
The genetic fidelity of micropropagated P. bipinnatifidum clones was confirmed by using PCR-based RAPD analysis. Polymorphism detected among these clones was negligible. The micropropagation protocol can successfully be employed for commercial application without any risk of genetic instability.
Author Contributions: Conceptualization, MSA, AAA and YHD; methodology, MSA and AAA; software, MSA and AAA; validation, YHD, DAS and MKS; formal analysis, MSA, AAA and YHD; investigation, MSA and AAA; resources, MSA and YHD; data curation, MSA and AAA; writing—original draft preparation, MSA, AAA and YHD; writing—review and editing, MSA, AAA, YHD; DAS and MKS; visualization, DAS and MKS; project administration, MSA and YHD; funding acquisition, MSA. All authors have read and agreed to the published version of the manuscript.
Funding: This research was funded by the Deanship of Scientific Research at King Saud University for funding this work through research group NO (RGP-1435-086).
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Data Availability Statement: All data are presented within the article.
Acknowledgments: The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group NO (RGP-1435-086).
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
REFERENCES
1. Dewir YH, Nurmansyah, Naidoo, Y, Teixeira da Silva, JA. Thidiazuron-induced abnormalities in plant tissue cultures. Plant Cell Reports. 2018. 37: 1451-1470.
2. Podwyszyńska M, Niedoba, K, Korbin, M, Marasek, A. Somaclonal variation in micropropagated tulips determined by phenotype and DNA markers. Acta Horticulturae. 2006. 714: 211–220.
3. Krishna H, Alizadeh, M, Singh, D, Singh, U, Chauhan, N, Eftekhari, M, Sadh, RK Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech. 2016. 6: 54.
4. Rai MK, Phulwaria, M, Harish, Gupta, AK, Shekhawat, NS, Jaiswal, U. Genetic homogeneity of guava plants derived from somatic embryogenesis using SSR and ISSR markers. Plant Cell, Tissue and Organ Culture. 2012. 111: 259-264.
5. Kanchanapoom K, Chunui, P, Kanchanapoom, K. Micropropagation of Anubias barteri var. nana from shoot tip culture and the analysis of ploidy stability. Notulae Botanicae Horti Agrobotanici Cluj-Napoca. 2012. 40: 148-151.
6. Sharma S, Sudheer Pamidimarri, D.V.N., Vijay Anand, KG, Reddy, MP. Assessment of genetic stability in micropropagules of Jatropha curcas genotypes by RAPD and AFLP analysis. Industrial Crops and Products. 2011. 34: 1003-1009.
7. El-Mahrouk ME, Dewir, YH, Naidoo, Y. Micropropagation and genetic fidelity of the regenerants of Aglaonema ‘valentine’ using randomly amplified polymorphic D.N.A. HortScience. 2016. 51: 398–402.
8. Dann AL, Wilson, CR. Comparative assessment of genetic and epigenetic variation among regenerants of potato (Solanum tuberosum) derived from long-term nodal tissue-culture and cell selection. Plant Cell Reports 2011. 30: 631–639.
9. Stimela T, Kasili, RW, Mamati, EG. Pomegranate micropropagation, callogenesis and genetic integrity assessment using simple sequence repeat markers. Journal of Agricultural Science 2019. 11: 237-250.
10. Kumar Mishra A, Nath Tiwari, K, Mishra, P, Kumar Tiwari, S, Kumar Mishra, S, Singh, J. Factors affecting the efficiency of in vitro regeneration from seedling-derived nodal explants of Nyctanthes arbortristis L. and evaluation of genetic fidelity. Plant Biosystems 2020. 154: 197-205.
11. Vemula S, Koppula, T, Jogam, P, Mohammed, M. In vitro high frequency multiplication and assessment of genetic fidelity of Corallocarpus epigaeus: An endangered medicinal plant. Vegetos. 2020. 33: 63-73.
12. Asthana P, Jaiswal, VS, Jaiswal, U. Micropropagation of Sapindus trifoliatus L. and assessment of genetic fidelity of micropropagated plants using RAPD analysis. Acta Physiologiae. 2011. 33:1821-1829.
13. Ahmad, N, Anis, M. An efficient in vitro process for recurrent production of cloned plants of Vitex negundo L. European Journal of Forest Research. 2011. 130: 135-144.
14. Ramesh, M, Vijayakumar, KP, Karthikeyan, A, Pandian, SK. RAPD based genetic stability analysis among micropropagated, synthetic seed derived and hardened plants of Bacopa monnieri (L.): a threatened Indian medicinal herb. Acta Physiologiae Plantarum 2011. 33: 163-171.
15. Alawaadh AA, Dewir, YH, Alwihibi, MS, Aldubai, AA, El-Hendawy, S, Naidoo, Y. Micropropagation of lacy tree Philodendron (Philodendron bipinnatifidum Schott ex Endl.). HortScience 2020. 55: 294-299.
16. Murashige T, Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologiae Plantarum 1962. 15: 473-497.
17. Doyle JJ, Doyle, JJ. Isolation of plant DNA from fresh tissue. Focus 1990. 12: 13-15.
18. Xiong ZJ. Regeneration of Philodendron Micans K.koch through protocorm-like bodies and improvement of plant form using growth regulators. MSc thesis, University of Florida, USA. 2009.
19. Gangopadhyay G, Bandyopadhyay, T, Gangopadhyay, SB, Mukherjee, KK. Luffa sponge - A unique matrix for tissue culture of philodendron. Current Science 2004. 86: 315-319.
20. David A, Madappa, MB, Bopaiah, AK. RAPD Analysis of rapidly multiplied in vitro plantlets of Anthurium andreanum Bicolour Var Agnihotri. IOSR-J.B.B. 2015. 1: 10-14.
21. Lal D, Singh, N. Mass multiplication of Celastrus paniculatus Willd an important medicinal plant under in vitro conditions using nodal segments. Journal of American Science. 2010. 6: 55-61.
22. Premvaranon P, Vearasilp, S, Thanapornpoong, S, Karladee, D, Gorinstein, S. In vitro studies to produce double haploid in Indica hybrid rice. Biologia 2011. 66: 1074–1081.
23. Bairu MW, Aremu, AO, van Staden, J. Somaclonal variation in plants: causes and detection methods. Journal of Plant Growth Regulation. 2011. 63: 147 – 173.
24. Samarfard S, Kadir, MA, Kadzimin, SB, Saud, HM, Ravanfar, SA, Danaee, M. In vitro propagation and detection of somaclonal variation in Phalaenopsis gigantea as affected by chitosan and thidiazuron combinations. HortScience 2014. 49: 82–88
25. Borsai O, Hârța, M, Szabo, K, Kelemen, CD, Andrecan, FA, Codrea, MM, Clapa, D Evaluation of genetic fidelity of in vitro-propagated blackberry plants using RAPD and SRAP molecular markers. Horticultural Science. 2020. 47: 21-27.
Received: 17 June2021 / Accepted: 20 August 2021 / Published: date. 15 February 2022
Citation: Alwahibi MS, Alawaadh AA, Dewir YH, Soliman DA, and Seliem MK. Assessment of genetic fidelity of lacy tree philodendron (Philodendron bipinnatifidum Schott ex Endl.) micro propagated plants. Revis Bionatura 2022;7(1). 10. http://dx.doi.org/10.21931/RB/2022.07.01.10
