2023.08.01.41
Files > Volume 8 > Vol 8 No 1 2023
Tumor necrosis factor (TNF)-α-308 gene polymorphism in children of Haemophilia A
1 University of Southern Technical, Institute of Al-Nasiriya Technical, Department of Nursing, Iraq
2 University of Southern Technical, Institute of Al-Nasiriya Technical, Department of Nursing, Iraq
3 University of Southern Technical, Institute of Al-Nasiriya Technical, Department of Nursing, Iraq
*Correspondence: [email protected] ; Tel.: 00 9647812451059
Available from: http://dx.doi.org/10.21931/RB/2023.08.01.41ABSTRACT
Hemophilias are the most common X-linked inherited blood diseases that, if not properly treated, can cause lifelong debilitations. The challenges and problems in babies differ from that in older kids and adults. Blood loss conditions continue to dominate as diagnostic triggers in children, but the locations of blood loss vary with age. The TNF-α-308 gene polymorphism in children with moderate to severe hemophilia correlates with genetic background and with the clinical phenotype of the cases. This study was a case control conducted in fifty hemophilic and fifty age- and sex-matched healthy cases from September 2020 to October 2021. Results; a significant change was found among positive and negative inhibitors regarding the number of factors eight exposure days >20 days (in positive inhibitors was 59.3% compared to adverse inhibitors 27.8%). Conclusion: The (TNF-)308 gene polymorphism is significantly correlated with inhibitor progress in severe Haemophilia A cases. TNF-Alpha gene might be of use as a biomarker as well possible immune response modulator in Haemophilia A patients receiving substitute treatment.
Keywords: polymorphism, TNF-α -308Haemophilia, inhibitor, prophylaxis - intracranial hemorrhage and pediatrics.
INTRODUCTION
Haemophilia A (HA) is the most everyday coagulation parameter shortage globally, with elevated treatment costs. It impacts all ethnic inhabitants, and its prevalence fluctuates between various states but is valued at rates of 3 to 20 cases/100k people 1. According to the World Federation of Hemophilia's (WFH) yearly report for 2016, the estimated total number of cases in Iraq with all types of hemophilia in 2016. There were 1346 cases, for a prevalence of 3.7/100,000 people 2. Genetically, HA is caused by omitting all or part of the factor VIII gene, a point mutation in the genes or gene regulation. Major inversions of the tip of the X chromosomes, which are adaptive for half of the severe degree of such diversity of abnormalities in the factor-VIII (FVIII) gene, cause a lack of reduced productions of the FVIII molecule, which has no role or decrease of FVIII in plasma 1. In severe hemophilia, phenotypic changeability is well-known.
Cases of HA have mutable clinical phenotypes that correlate with FVIII plasma levels and other genetic and environmental influences. The precise cause of this clinical heterogeneity remains unknown. The FVIII gene mutation, on the other hand, is regarded as the most critical determinant of the clinical phenotype 3. TNF is considered a cytokine with multiple functions. TNF- is of an essential role in defense for the host in contradiction of infections and has a primary function in auto-immune disorders. It is also a necessary cytokine for the formation of granulomas. TNF-alpha levels vary from case to case and are genetically determined. A gene encodes TNF-alpha on 6p21, part of the main histocompatibility complex (MHC). The third chromosome was a highly polymorphic region. Several bi-allelic as (SNPs) were found rounding (in or and) the gene of TNF-α. One G/A polymorphism is situated at the gene -308 upstream and is identified to impact the TNF level. Compared to the allele as TNF—308G, allele A has more excellent transcription activities 4.
We wanted to look into TNF—308 gene polymorphism in children with moderate to severe Haemophilia and see if there was a link between genetics and clinical phenotypes
MATERIALS AND METHODS
Cases and Methods
This study was a case-control study performed from September 2020 to October 2021 for fifty hemophilic cases& fifty healthy cases of matched age & sex attending hematology at Al-Hussein teaching hospital in Nasiriya.
Inclusion criteria: Age group: > 1 month to18 years old, moderate and severe hemophilic cases, Haemophilia A was sorted regarding the level of serum-FVIII: <1 % of ordinary (severe), 1–5% of regular (moderate), and >5%of every day (mild) and accept to sign the informed consent form
Exclusion criteria: Rather than Haemophilia and associated chronic illness, any associated hematologic disorders.
All cases will be subjected to the following
A- Full history taking including ages, gender, the existence of parental consanguinities, ages at onset of cyanosis, cyanotic conditions and their incidences, blood loss manifestation, thrombocytopenia family history, medication past, family history of Hemophilia, FVIII activity, FVIII brand, ages of first exposures to FVIII.
B- Complete clinical examination: Physical appearance with specific attention to abnormalities of growth, signs of anemia, arterial blood pressure and other vital signs and data such as temp, heart & respiratory rates and BP have been determined.
C- Investigations: 2 ml of venous blood were gathered from all cases and control groups, placed in tubes containing EDTA, and reserved at -20 °C until DNA extraction. DNA was extracted using commercial DNA extracting kits and kept at -20°C.
D- Standard approaches: Southerly blot and wide-range chain reactions (PCR) have been used to analyze the FVIII gene in versional analyzing, PCR and mutation screening approaches.
E- TNFA polymorphisms: The isolated DNA for the polymorphic locations-308 was genotyped for TNF-polymorphism using polymerase chain reactions (PCR) and a thermal cycler. Polymorphisms were studied using the (RFLP) technique and to sites -308 primers specific. The TNF-alpha-308G/A (rs-1800629) polymorphism was discovered using a primer as single, which produces a place as NcoI recognition in the allele as A; therefore, PCR (107 bp) absorption with yields of NcoI being two results (87 and 20 bp). The G-allele is still present. TNF-A amplifications were carried out using a thermal cycler. From blood EDTA-conserved, TNF-A polymorphisms g-DNA was extracted utilizing (Qiagen-genomic tip, Kebo-Lab, Sweden) Kit. The polymorphism as TNFA 308GA (rs1800629) was discovered using primer being single, which generates a site of NcoI-recognizing in the allele as A; therefore, the PCR ingestion (107 bp) along with NcoI (Promega, Germany) yields two results (87 and 20 bp). The G-allele is still present. A thermal cycler (Techne, PHC3 Dri block, and Cambridge, UK) achieved TNF-A amplification amplifications. For 3.5 hours, the digested NcoI samples were exposed to agarose gel electrophoresis in three percent Nu-Sieve GTG at 5-V per cm.
F- Procedure: Extractions of DNA and polymerase chain reactions: DNA was extracted utilizing Gene JET as Total Blood DNA Genomic Purifications Mini Kits (Qiagen genomic tip, Kebo Lab, Stockholm, Sweden) and kept at -20 °C. The primer sequence used is depicted in (Table 1). The reaction volume was 25l: 5l of DNA at 100ngl, 15.0 l Dream Taq Green PCR master mix (thermo-scientific, USA), 0.5 l of every primer (25pmoll), and 4.0l H2O. The reaction was carried out in a thermocycler TC-100 (Biorad, USA) along with the following parameters as cycling: rs1800629: The profile of cycling was as follows: 1st denaturation at 940 degrees Celsius for three minutes, thirty cycles of 94 degrees Celsius for 10 seconds, and appropriate annealing temp. Sixty degrees Celsius for 30 seconds, 72 degrees Celsius for 30 seconds, and ultimate extensions for 5 minutes at 72 degrees Celsius. For checking the products of PCR at 107 bp for rs1800629, ten l of PCR were resolute in agarose gel 2 percent.
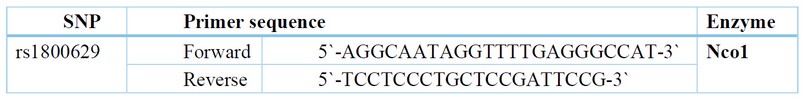
Table 1. Primer sequence and PCR-RFLP enzyme
DNA concentration, purity and purity: were measured spectro-photometrically utilizing the Thermo Scientific NanoDropTM Spectro-photometer (Nanodrop). Following the formulations: DNA concentration (g per ml) = (A-260–A-320 readings) dilution factor 50g per ml DNA yield (g) = total specimen volume x DNA concentration (ml). The re-activity of every specimen was determined by dividing the average specimen OD by the low positive controls average OD. The final result was multiplied by the number of units assigned to the low positive. After that, samples were labeled being negative (less than 20 units), moderately positive (from 20 to 60 teams) or strongly positive (greater than 60 units).
Statistical analysis: Statistical analysis has been done by SPSS-21 (IBM, USA). The normality of data has been first examined with one-sample Kolmogorov-Smirnov testing. Associations among qualitative variables were done via Chi-square testing, whereas Fischer exact testing and Monte Carlo testing were employed when the expected cell count <5. Comparison between both groups was made by means of Student t-testing for parametric results and Mann-Whitney testing for nonparametric results.
RESULTS
As regards characteristics of studied cases, most of them presented with spontaneous bleeding due to minor trauma (73.3 percent ). In addition, most patients (84.4 percent) had negative consanguinity and no Haemophilia family history (55.6 percent ). Also, the number of factor exposure days < 20 represents 53.3% of cases. The number of bleeding episodes per month was < one time per month in 57.8%. No first exposure to factor 8 was 55.6% in 25 cases with age < 18 months. All patients in the study were given recombinant factor 8 concentrate therapy on demand at an average dose of 30 iu/kg. 26 of hemophilic cases (57.8%) had factored 8 activities less than 1%. Moderate Haemophilia represents 60% of cases; severe Haemophilia represents 40%. Table (2)
A non-significant change was found among the studied group regarding genotype, but GA distribution was higher in the patient group (15.6%) than in the control group (6.7%). Regarding alleles, the A-allele distribution was higher in the patient group (7.8%) compared to the control group (3.3%). Table (3)
There is no significant difference in positive family history between moderate and severe Haemophilia, but the frequency was higher in severe Haemophilia (61.1%) than in moderate Haemophilia (33.3%). There is a significant difference between average and severe Haemophilia as regards several intensive factors 8 treatment; the frequency was higher in severe Haemophilia (median =8) than in moderate Haemophilia (median =5). There is a significant difference between average and extreme Haemophilia as regards positive consanguinity; frequency was higher in the middle than in severe Haemophilia. Table (4)
A non-significant change was found among moderate and severe Haemophilia regarding genotype, but GA distribution was higher in the middle than severe Haemophilia. Also, the A allele frequency was higher in moderate than powerful Haemophilia. Table (5)
A non-significant change was found among positive and negative inhibitors regarding genotype, but GA distribution was higher in positive inhibitors 22.2% than in adverse inhibitors 5.6%. Also, A allele distribution was increased in positive inhibitors at 11.1%, then in negative inhibitors at 2.8%. Table (6)
Following logistic regression analysis and adjusting for confounding factors, the only independent predictor of positive inhibitors was the number of factors 8 exposure days > 20 days (OR = 3.8) with a 95% CI (1.05-13.7).Table (7).
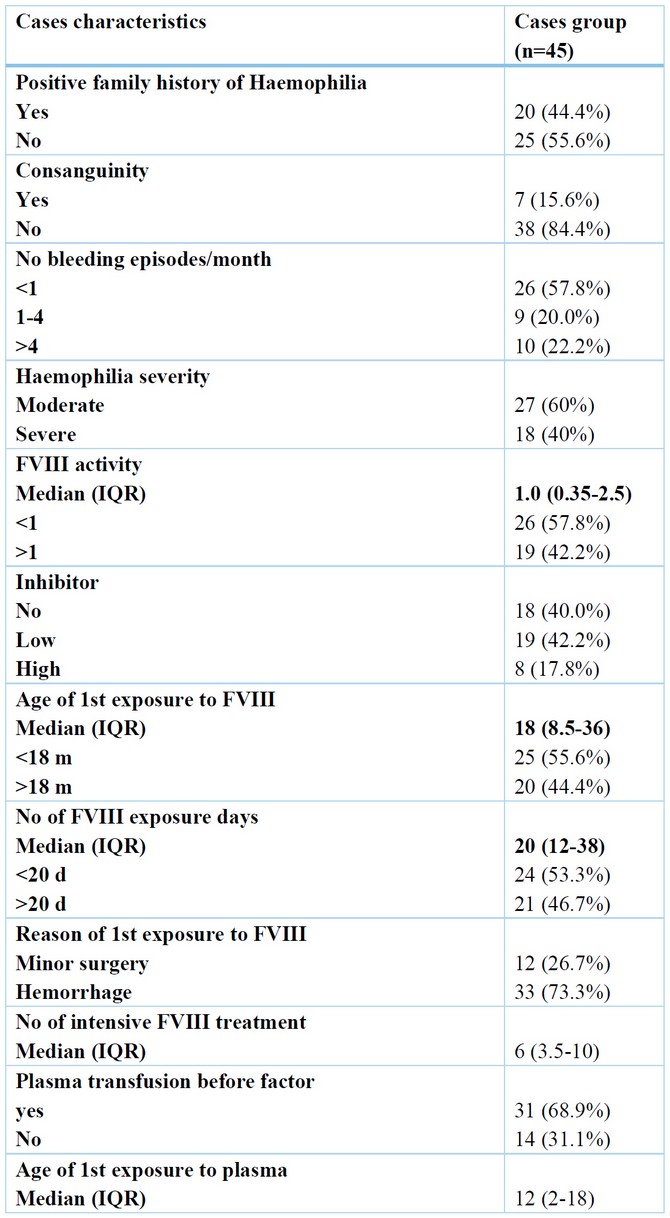
Table 2. Cases characteristics regarding disease
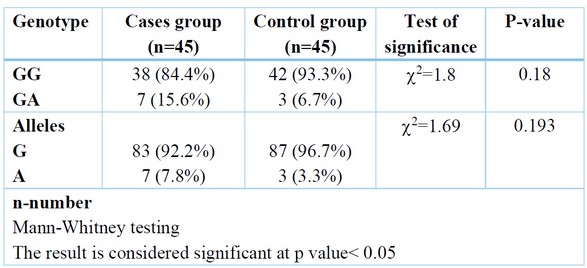
Table 3. Comparing of Genotype among cases and control groups.
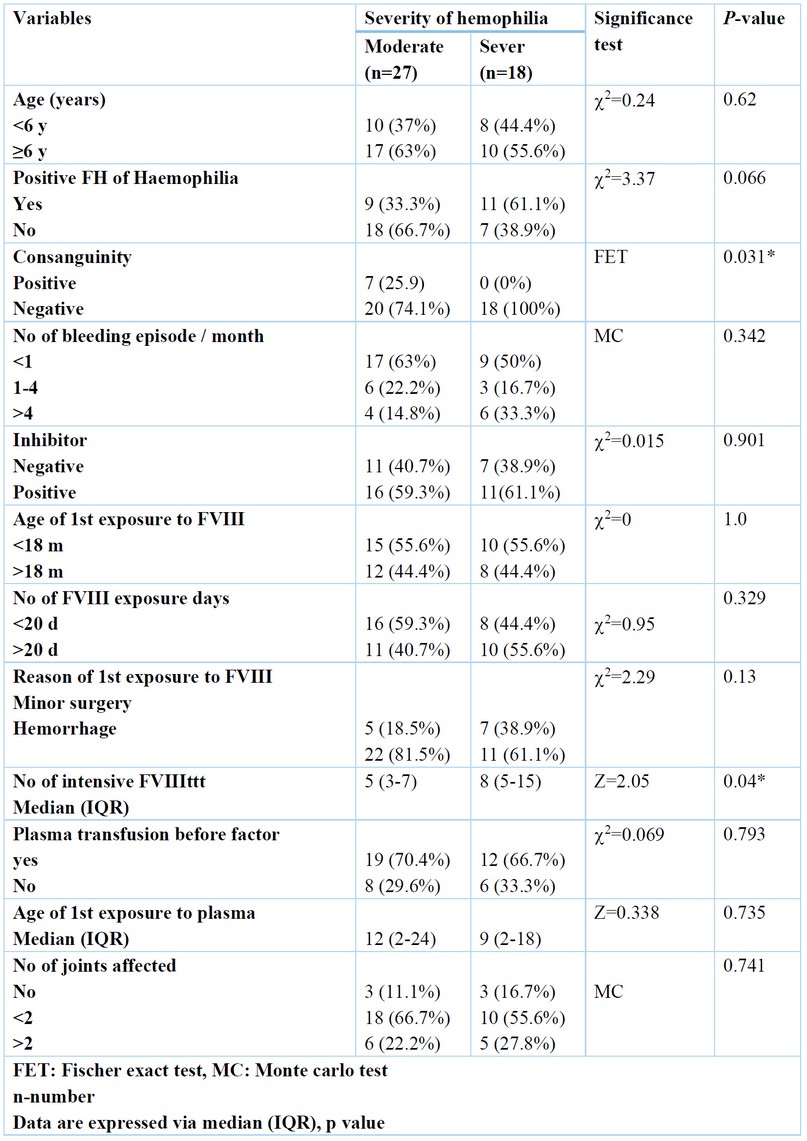
Table 4. Relation between Haemophilia severity and patient's characteristics
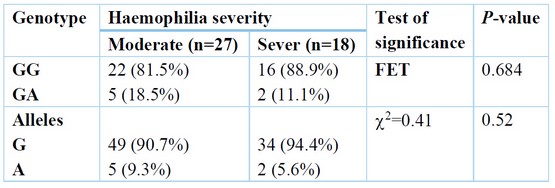
Table 5. Relation between Haemophilia severity and genotype.
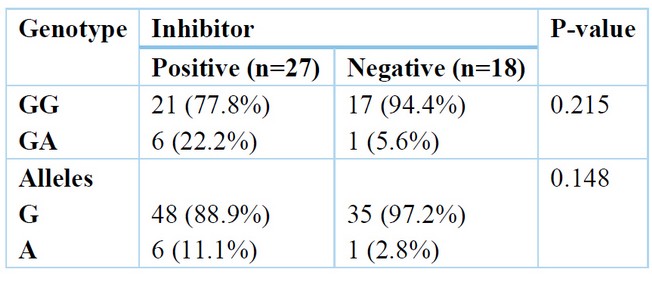
Table 6. Relation between inhibitor and genotype.
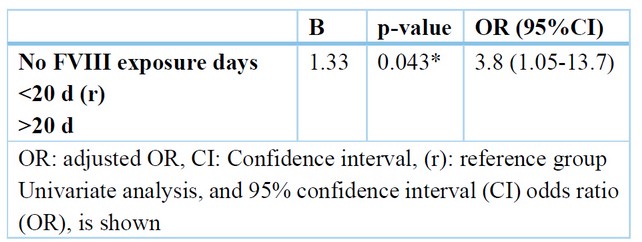
Table 7. Relation between duration for taking OCS and gingival index.
DISCUSSION
TNF—308 promoter gene polymorphism was linked to various auto-immune diseases, counting systemic rheumatoid arthritis, lupus erythematosus, and infections like TB. No studies on TNF- gene polymorphisms in TA were discovered throughout the study writing process. During the manuscript review process, we came across one report that found no TNF- promoter polymorphism in the Han Chinese population. As a result, they conducted this pilot report to investigate TNF—308 polymorphism in TA, a granulomatous vasculitis with an auto-immune basis that is most likely caused by Mycobacterium tuberculosis 5. This is why this study was chosen to investigate (TNF)—308 gene polymorphisms in Iraqi children with moderate to severe hemophilia and to correlate the genetic background with the clinical phenotypes of the cases. In Iraq, a single Hereditary Blood Disorders Center in every governorate delivers care for these cases, excluding Baghdad, where three centers exist. A case-control study was conducted comprising 50 hemophilic cases& 50 healthy cases of matched age & sex. Total 45 hemophilic cases and 45 controls were screened for participation during this study. There is a non-significant change among the study groups regarding age and sex. Regarding age, the cases group had a median 6 IQR between 5-10 compared to the control group, which had a median 5 IQR between 3-11. Regarding sex, the majority of both groups were males. Our results are supported by the study of Nesheli et al., 6,7 as they reported that the mean age of cases with inhibitors was 10.22 years. Their results revealed that the production of inhibitors in young populations was more significant than in older ones. It might be because of the usage of exogenous FVIII as prophylaxis in some young Haemophilia A cases and the deficiency of use of any factor on demand in the old cases 8.9. However, Astermark et al., 10 reported that the median age of the studied group was 24 (3-79)-yrs, and all cases had >100 exposures to FVIII concentrates. The present study shows that as regards characteristics of studied patients. Most of them presented with spontaneous bleeding due to minor trauma (73.3 percent ). Furthermore, most cases (84.4 percent) had negative consanguinity and no Haemophilia family history (55.6 percent ). Also, the number of factor exposure days < 20 represents 53.3% of cases. The number of bleeding episodes per month was < one time per month in 57.8%. No first exposure to factor 8 was 55.6% in 25 cases with age < 18 months. All cases in the study received recombinant factor 8 concentrate therapy on demand at an average dose of 30IU/kg. Twenty-six hemophilic cases (57.8%) had factor 8 activity less than 1%. Moderate Haemophilia represents 60% of cases; severe Haemophilia represents 40%. Taresh et al., 11 who conducted their study in Basra, Iraq, 39.8-% of enrolled cases with HA had a severe form of the disorder, and more than 33% were >15 years old, while 56% of the cases with HB had a mild grade of the disease, and nearly half were younger than five years old. Our results are supported by the study of Al Tonbary et al., 12 as they reported that moderate presentation represents the majority in 17.2%, followed by severe presentation in 4.7%. The mean presentation symptoms were hemorrhage followed by males' circumcisions at 51.4%, trailed by posttraumatic bleeding at 36.1%. Unplanned detection was revealed to be shared in females.
However, Astermark et al., 10 reported that number of 124 cases, 75.6% have severe (52 cases with HR, 11 cases with LR, 61 patients with no inhibitors), 15.9% were moderate (2 cases with HR, 5 cases with LR, 19 cases with no inhibitors), and 8.5% mild (3cases with HR, 4cases with LR, 7cases with no inhibitor) HA. In the study in our hands, non-significant changes were found among the study groups regarding genotype. However, GA distribution was higher in-patient group (15.6%) than in the control group (6.7%). Regarding alleles, the distribution of the A-allele was high in the patient group (7.8%) compared to the control group (3.3%). Our results are supported by the study of Zhang et al., 13 as they reported that in cases of HA, genotypes as G/A, G/G, and A/A have been found in 12.8%, 84.3% and 2.9% resp.; C/C, C/T and T/T genotypes have been found in 77.2%, 21.4%, and 1.4 %resp. The variance in the genotype incidences among the study groups was non-significant.
Furthermore, Sandhya et al., 14 Three results were obtained after enzyme digestion of the PCR products. A comprehensive NcoI cut for homozygous TNF- (-308G/G), yielding 2 87 and 20 base pair remains; an incomplete cut for heterozygous TNF- (-308G/A), producing 3 107, 87, and 20 base pair remains; and an un-cut homozygous TNF- (-308A/A), yielding 107 base pair fragments. The difference in genotype distribution of the polymorphism between the two groups was not statistically significant. The present work shows that there is non-significant change among moderate and severe Haemophilia regarding genotype, but GA distribution was higher in moderate than severe Haemophilia. Also, A allele frequency was higher in moderate than severe Haemophilia. The study of Astermark et al. supports our results, 10 They reported that for the current work, Hap 2 (Hap 2-2) homozygous was of significantly greater hazard of inhibitor progression compared to Hap 1-1, and the single variance between such two haplotypes is the uncommon allele being TNFA–308A, which has been linked to pathophysiologic roles in definite antibody-mediated auto-immune disorders. The correlation along with such allele was stronger in the sub-group of cases with HA (OR, 19.2; P value.001) being severe, and it was as well found in the collection of seventy-five cases with overturns—which is, in a group of cases with a similar hazard for inhibitors emergence according to the causal FVIII mutations themselves. This correlation was more than non-dependent on allele-134 of IL-10G 15. The current work shows that a non-significant change was found among positive and negative inhibitors regarding genotype, but GA distribution was higher in positive inhibitors 22.2% than in negative inhibitors 5.6%. Also, A allele distribution was more increased in positive inhibitors, 11.1%, than in negative inhibitors, 2.8%. Following logistic regression analysis and adjusting for confounding factors, the only independent predictor of positive inhibitors was the number of factors 8 exposure days > 20 days (OR = 3.8) with a 95% CI (1.05-13.7). Zhang et al. 13 reported that cases with homo-zygotes for A-allele had an advanced danger of inhibitors advance compared to those who weren't (OR = 7.519, 95% CI = 3.168 - 17.844). Severe HA cases with homo-zygotes for A-allele had an elevated danger of inhibitors advancing compared to those who weren't (OR = 8.163, 95% CI = 2.521 - 26.434). A non significant change in the danger of inhibitors advance among the cases having or not (OR = 1.586, 95% CI = 0.729 - 3.450). This can be revealing of a causal association of TNFα –308 A/A with inhibitors formations and the elevated incidence connected to the choice of inhibitors cases (47.0%) for the report in the group of MIBS, or to other inherited influences alongside Hap-2 But, the opportunity in which the 308A-allele is additionally predominant in Haemophilia cases with inhibitor compared to such without inhibitor and in controls requisites to be assessed in multi-ethnic populations-built groups. The inhibitory progression of antibodies in response to exogenous FVIII is frequently measured as a Th2-cell-induced immune response, while TNF- is chiefly associated with Th1 cells. But, the cytokine profile obviously shows that inhibitor formations are a mixed Th1 and Th2 cell responding, emphasizing that TNF- levels can similarly moderate the immune response to missing factors in Haemophilia cases. (1). In a study by Wilson et al., 17, The HLA B8, A1, and DR3 alleles were significantly correlated with 17 TNFA –308A. However, while our findings strengthen a link among the polymorphism –308A and such alleles, no one was additionally common in inhibitors cases in their group, with only B44 being the enriched allele of an OR of 2.1—which is analogous to what is defined through Oldenburg et al., 18 (34.5% vs. 16.7%; OR, 2.2).
CONCLUSIONS
In cases of severe Haemophilia A, TNFα-308 gene polymorphism is significantly correlated with inhibitor progress. TNF- gene can be a beneficial bio-marker and potential modulator of immune responding to auxiliary therapy in Haemophilia A cases.
Author Contributions: Thaer Ali Hussein: Conceptualization, Writing – original draft. Ali A.H. AL-bakaa: Visualization, Methodology, Project administration. Mohammed Hassan Flaih: Validation, Resources.
Funding: This research received no external funding.
REFERENCES
1. Mansouritorghabeh H. Clinical and laboratory approaches to hemophilia A. Iranian journal of medical sciences. 2015 May;40(3):194.
2. Tonbary YA, Elashry R. Descriptive epidemiology of hemophilia and other coagulation disorders in mansoura, egypt: retrospective analysis. Mediterranean journal of hematology and infectious diseases. 2010 Aug 13;2(3):e2010025-.
3. Carcao MD, et.al. Correlation between phenotype and genotype in a large unselected cohort of children with severe hemophilia A. Blood, The Journal of the American Society of Hematology. 2013 May 9;121(19):3946-52.
4. Astermark J, et.al. MIBS Study Group. Polymorphisms in the TNFA gene and the risk of inhibitor development in patients with hemophilia A. Blood. 2006 Dec 1;108(12):3739-45.
5. Lv N, et.al. The role of tumor necrosis factor-α promoter genetic variation in Takayasu arteritis susceptibility and medical treatment. The Journal of rheumatology. 2011 Dec 1;38(12):2602-7.
6. Nesheli HM, Hadizadeh A, Bijani A. Evaluation of inhibitor antibody in hemophiliaA population. Caspian journal of internal medicine. 2013;4(3):727.
7. SAl-Shaheeb, H Kamil Hashim, A Kadhim Mohammed, H Abdulkareem Almashhadani, A Al Fandi. Assessment of lipid profile with HbA1c in type 2 diabetic Iraqi patients. Revis Bionatura 2022;7(3) 29.
8. Franchini M, Tagliaferri A, Mengoli C, Cruciani M. Cumulative inhibitor incidence in previously untreated patients with severe hemophilia A treated with plasma-derived versus recombinant factor VIII concentrates: a critical systematic review. Critical reviews in oncology/hematology. 2012 Jan 1;81(1):82-93.
9. Al Gburi RH, Hashim RD, Kadhim HA, Adam Ş, Almashhadani HA. Correlation of Angiotensin-converting enzyme inhibitors and angiotensin II receptor blockers with serum GDF-15 in a group of hypertensive Iraqi patients. 2022 Aug 30;52(13.96):13-96.
10. Astermark J, Oldenburg J, Carlson J, Pavlova A, Kavakli K, Berntorp E, Lefvert AK, MIBS Study Group. Polymorphisms in the TNFA gene and the risk of inhibitor development in patients with hemophilia A. Blood. 2006 Dec 1;108(12):3739-45.
11. Taresh AK, Hassan MK. Inhibitors among patients with hemophilia in Basra, Iraq–A single center experience. Nigerian journal of clinical practice. 2019 Mar 1;22(3):416-.
12. Tonbary YA, Elashry R. Descriptive epidemiology of hemophilia and other coagulation disorders in mansoura, egypt: retrospective analysis. Mediterranean journal of hematology and infectious diseases. 2010 Aug 13;2(3):e2010025-.
13. Zhang LL, Yu ZQ, Zhang W, Cao LJ, Su J, Bai X, Ruan CG. Relationship between factor VIII inhibitor development and polymorphisms of TNFα and CTLA-4 gene in Chinese Han patients with hemophilia A. Zhonghua xue ye xue za zhi= Zhonghua Xueyexue Zazhi. 2011 Mar 1;32(3):168-72.
14. Sandhya P, Danda S, Danda D, Lonarkar S, Luke SS, Sinha S, Joseph G. Tumour necrosis factor (TNF)-α-308 gene polymorphism in Indian patients with Takayasu's arteritis-A pilot study. The Indian journal of medical research. 2013 Apr;137(4):749.
15. Astermark J, Oldenburg J, Pavlova A, Berntorp E, Lefvert AK, MIBS Study Group. Polymorphisms in the IL10 but not in the IL1beta and IL4 genes are associated with inhibitor development in patients with hemophilia A. Blood. 2006 Apr 15;107(8):3167-72.
16. Reding MT, Lei S, Lei H, Green D, Gill J, Conti-Fine BM. Distribution of Th1-and Th2-induced anti-factor VIII IgG subclasses in congenital and acquired hemophilia patients. Thrombosis and haemostasis. 2002;88(10):568-75.
17. Wilson AG, De Vries N, Pociot FD, Di Giovine FS, Van der Putte LB, Duff GW. An allelic polymorphism within the human tumor necrosis factor alpha promoter region is strongly associated with HLA A1, B8, and DR3 alleles. The Journal of experimental medicine. 1993 Feb 1;177(2):557-60.
18. Oldenburg J, Picard JK, Schwaab R, Brackmann HH, Tuddenham EG, Simpson E. HLA genotype of patients with severe haemophilia A due to intron 22 inversion with and without inhibitors of factor VIII. Thrombosis and haemostasis. 1997;77(02):238-42.
Received: December 23, 2022 / Accepted: January 30, 2023 / Published:15 February 2023
Citation: Ali Hussein T, AL-bakaa A A H, Hassan Flaih M. Tumor necrosis factor (TNF)-α-308 gene polymorphism in children of Haemophilia A. Revis Bionatura 2023;8 (1)41. http://dx.doi.org/10.21931/RB/2023.08.01.41
