2023.08.04.31
Files > Volume 8 > Vol 8 no 4 2023
Diversity and enzymatic activity of some fungi isolated from agricultural soil
Nemat A. Muhsen1* and
Mustafa A. Al-Dossary1
1University
of Basrah, College of Science, Department of Ecology
*Corresponding
author:email: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.04.31
ABSTRACT
Fungi are one of the most important groups of
microorganisms in the environment, and due to their ability to produce several
types of enzymes, they play an essential role in the environment. During this
study, fourteen species of fungi were isolated from fifteen soil samples collected from
several agricultural areas in Basrah Governorate, southern Iraq, and their
enzymatic activity was tested for four extracellular enzymes (cellulase C,
laccase La, lipase Lp, and manganese peroxidase Mp) on specific solid media.
The isolated fungi showed good enzymatic activity, in which 12 fungal species
can secret manganese peroxidase, 11 can secret cellulase, 9 can secret lipase,
and five can secret laccase enzyme. Aspergillus candidus and A. versicolor showed a positive
detection for all enzymes, Cladosporium showed positive detection for C,
Lp, and Mp, while Mucor sp. showed negative detection for all enzymes.
Keywords: Enzymatic activity, Fungal diversity, soil.
INTRODUCTION
Fungi
play an essential role in the decomposition process of organic substances by
enzyme secretion, such as (cellulase, laccase, lipase, and manganese
peroxidase). The microbes need to produce an extracellular enzyme to convert
polymeric compounds such as cellulose, lignin, starch, pectin, and other
components into smaller molecules that can be assimilated easily 1.
Many
researchers have tended to exploit these compounds to produce simple
saccharides of industrial importance, such as biofuel
production, beverage, confectionery, textile,
and leather. They are also used in the biological treatment of organic and
inorganic pollutants using the enzymes produced by fungi. There are different
types of fungal enzymes involved in this process, such as cellulase enzyme, which is capable of
decomposing cellulose into glucose 2; the laccase enzyme can break
the lignin that gives the wood its added firmness additionally,
it breaks down aromatic
hydrocarbon molecules 3, lipase enzyme stimulates several chemical
reactions for the hydrolysis of fats, which are crucial components of
agricultural and oil waste 4, and manganese peroxidase enzyme which is secreted mainly by white rot fungi and play a
dynamic role in the polymerization and depolymerization of lignin and the
oxidation of phenolic and non-phenolic compounds 5.
The
degradation of all organic and agricultural substances depends on the presence
or the absence of enzymes secreted by the microorganisms
and the strength of the enzymes themselves
6. These extracellular enzyme systems secreted into their surrounding environment
allow the fungi to grow on various natural and artificial substrates, where
they break down a variety of substrates into small molecules that may be taken
up and digested by their cells 7.
The present study investigated the
diversity of fungi in some agricultural soil and evaluated their ability to
secrete cellulase, lipase, laccase and manganese peroxidase enzymes.
MATERIALS AND METHODS
Sampling
sites
Fifteen
soil samples were collected from various agricultural locations in the Basrah
Governorate. These areas include Abu Al-Kasib, Garmah Ali and Alqurna. These locations
have different plant types ranging from date palms to vegetables. 250 g of
agricultural soil was taken from the soil each time. Each sample was taken from
other locations and mixed to form one homogenized sample.
Isolation and identification of fungi
Dilution plate method 8,
was used to isolate fungi from fifteen soil samples collected from various
agricultural locations in the Basrah Governorate, in which 10 g of soil were
diluted in 90 ml Distilled water to make a dilution of 10-1. Potato
dextrose agar medium (PDA) supplemented with 250 mg/l
chloramphenicol antibiotic was used for the cultivation and isolation of
fungi from the diluted soil samples; it was prepared according to the direction
of the manufacturing company (Hi-Media, India). The cultures were incubated at
25 °C and examined first after 3-4 days from incubation to see the fungal
hyphae, and they were further set for one to two weeks.
The isolated fungi were first tested
under the dissecting microscope. Then, slides were prepared from the isolated
fungi and stained with lactophenol cotton blue dye to see the microscopic
features of each fungal isolate under the compound microscope. The isolated
fungi were identified according to the following 9, 10, 11, 12, 13.
The
percentage of occurrence for the isolated fungi was recorded according to the
following equation:

Evaluation of the enzymatic
activities of the isolated fungi
The enzymatic
activity was evaluated using special media; pure cultures from all fungal
isolates were first activated on a PDA medium for one week at 25 °C. Then, a
disk was taken by a cork borer 5 mm from the edge of each fungal isolate and
used to inoculate the center of the media to study the enzymatic activity for
each enzyme as
follows.
Cellulase
enzyme
Carboxymethyl cellulose medium (CMC)
was used to evaluate the ability of the isolated fungi to secret cellulase
enzyme; this medium contained g/l: 1g K2PO4, 0.5g KCL, 2g
NaNo3, 0.5g MgSO4: 7H2O, 2g carboxy methyl cellulose, 20g agar and 1L distilled water 14. After 3-7 days
of incubation at 25 °C, the plates were flooded with 0.2% aqueous Congo red
solution and distained with 1M NaCl for 15 minutes. The appearance of yellow
areas around the fungal colony indicates positive activity; otherwise, the red
medium shows harmful activity. 1.
Lipase
enzyme
Peptone
agar medium was used to evaluate the ability of the isolated fungi to secret
lipase enzyme; the composition of this medium was g/l: 10g peptone, 5g NaCl,
0.1g CaCl.2H2O, 20g agar and 1L distilled water; pH6.0
supplemented with 1% Tween 20 separately sterilized by filtration using
Millipore filter paper 0.45 µm and added to the medium. After seven days of
incubation at 25 °C it was observed that some fungal isolates formed visible
precipitation around the colony due to the formation of calcium salts of the
lauric acid liberated by the enzyme, which indicated positive lipase activity of
the fungi, while others didn’t form any precipitation which means hostile
activity 1.
Laccase
enzyme
The
ability of fungi to secret laccase enzyme was evaluated by using Glucose yeast
extract peptone agar medium containing g/l: 20g glucose, 5g yeast extract,10g
peptone, 20g agar and 1L distilled water and supplemented with 0.05g
α-naphthol, pH 6.0. After seven days of incubation at 25 °C, it was observed
that only the fungal isolate, which has a positive ability to secrete laccase
enzyme, turns the colorless medium into dark due to the oxidation of α
-naphthol by laccase enzyme 1.
Manganese
peroxidase enzyme
The
ability of fungi to secret manganese peroxidase enzyme was evaluated by using
Czapek-dox agar medium, which was prepared according to the
direction of the manufacturing company (Hi-Media, India), and then supplemented with phenol red dye
at a concentration of 0.0025%. After seven days of incubation at 25 °C, it was
observed that some fungal isolates turned the color of the medium from red to
yellow, which indicates a positive reaction, and the isolate can secret
manganese peroxidase enzyme; otherwise, red medium indicates negative activity 15.
Statistical
analysis
The
ANOVA analysis was used by applying Minitab ver.16 to statistically analyze the
results of the fungal enzymatic activity. The mean was tested using the least
significant difference RLSD test under the probability level 0.01.
RESULTS AND DISCUSSION
Fungal identification
Fourteen
isolates were counted from fifteen agricultural soil samples (table 1). These
belong to 8 fungal genera in addition to sterile mycelia. Thirteen species from
them, with 92.85 % percentage of occurrence, belonged to ascomycota either in
their anamorphic state with 12 species or with its telemorphic state with only
one species, Chaetomium sp. in the second place came the
zygomycota with only one species Mucor sp. and 7.1 % percentage of
occurrence.
The appearance of the ascomycetes fungi with
their anamorphic state in high percentage was due to their ability to produce
many reproductive units. This allows them to spread quickly in the environment.
Also, they can secrete different types of enzymes, enabling them to use other
materials in the environment for growth. Besides, they can tolerate the stress
in the atmosphere; these features allow them to be one of the most widespread
groups of fungi in the environment 16, 17, 18, 19.
The number of isolated fungi in this study
seems to be low compared to the other studies; this may be due to the high
temperatures during the time of sample collection, which may reach 50 °C, which
negatively affects the growth of fungi in the environment. In general, the
differences in the percentages of appearance of fungal genera may be due to
their ability to tolerate and adapt to extreme conditions, their adaptation to
a wide range of temperatures and their ability to secrete different types of
enzymes that enable them to decompose various materials and exploit them as a
source of energy and growth, in addition to their ability to produce a large
number of reproductive units which enable them to spread in the environment 20.
The percentage of occurrence of
species ranged from 7.1%, like Alternaria
sp. and Bipolaris sp., to 57.1 % in Aspergillus
niger; most of the isolated species belonged to the genus Aspergillus
with 7 species. This genus has high enzymatic activity and produces large
quantities of reproductive units,
which enable its species to adapt very well to their environment. It can grow
and widely spread in the environment 12.
This result is consistent with
other studies in which the Ascomycetes and the genus Aspergillus
represent the most isolated fungi 21,22,23, 24.
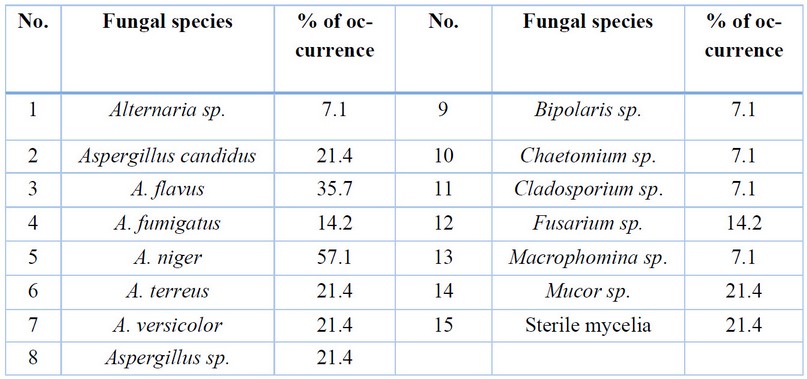
Table 1. The isolated fungi with
their percentage of occurrence
Enzymatic
activity
The enzymatic activity of 14 fungal species
isolated from agricultural soil was evaluated to study their ability to secrete
cellulase, laccase, lipase and manganese peroxidase enzymes.
However, there is an evident variation
between the different fungal isolates in terms of their enzymatic abilities;
the results showed that there were significant differences between the other
fungal species in their enzymatic activity, whether between the species
belonging to one genus or between species of different genera in terms of the
number of enzymes that each fungus was able to secrete and the quantity of its
secretion. The isolates showed diverse levels of enzymatic activity, and this
may be due to the enzymatic capacity that differs from one species to another
according to its adaptation to the environment in which it lives and the
inherent enzymatic activity of each fungus, in fact, in the atmosphere each
fungal species possesses an enzymatic capacity that distinguishes it from
other. 1, 25
The results showed that the tested
fungi could secrete from one to four different types of examined enzymes except
the Mucor sp., which was unable to secrete any enzymes. In comparison,
the species Aspergillus candidus and A. versicolor were able to
secrete all four types of enzymes but in varying ability.
In general, when some species appear to have a negative result,
this doesn’t mean they don’t have any enzymatic activity. It may refer to
either it producing an enzyme but doesn’t liberate from the hyphae or it
produces and liberates. Still, the medium limits enzyme secretion; therefore,
the negative results do not represent an absolute confirmation of the species’
inability to make the specific enzyme 26.
Most
of the studied fungi were able to secrete manganese peroxidase enzyme, and 12
species of fungi were able to secret it, but with different capabilities; it gave a positive reaction by
changing the color of the medium from red to yellow due to the transformation
of phenol aromatic ring. The significant change in color of the media refers to the greater
secretion of this enzyme by the fungus with different capabilities of secretion
rates from low as in Alternaria sp., to high secretion as in Aspergillus
candidus (Table 2, Fig.1).
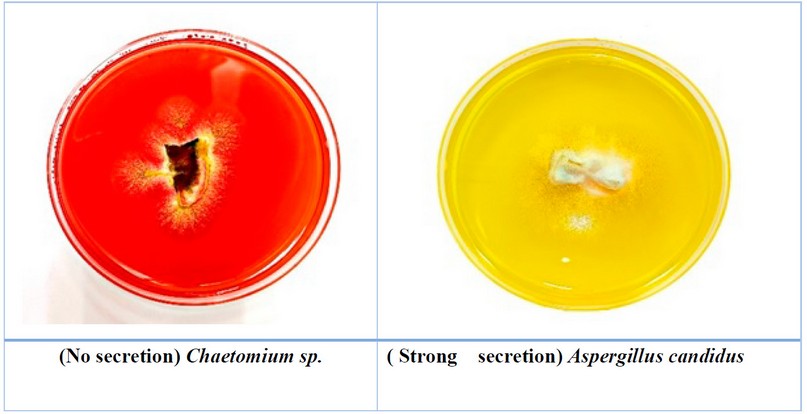
Figure 1. The ability of some fungi to secret manganese peroxidase
The
fungi that can secrete this enzyme are distinguished by their ability to
decompose complex pollutants and convert them into substances used as a source
of energy and carbon25.
Numerous
fungi possess the manganese peroxidase enzyme, a part of the ligninolytic,
extracellular enzymatic system primarily responsible for degrading lignin. It
can also degrade organic pollutants and is a commonly used enzyme in converting
toxic environmental contaminants into less toxic ones. Therefore, the manganese
peroxidase enzyme plays an essential role in the biological activity of fungi
due to its ecological importance and fungi that can secrete this enzyme are
distinguished by their ability to decompose complex pollutants and convert them
into substances used as a source of energy and carbon
27. This result is consistent with the study of 28.
who has found that most of his fungal isolates could secrete this enzyme.
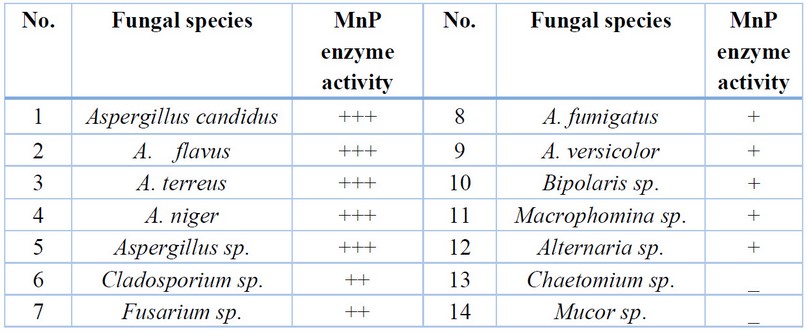 _
_- No color change, + Simple
color change, ++ Medium color change, +++ Strong color change
Table 2. Fungal enzymatic activity for manganese
peroxidase enzyme
The cellulase enzyme came in
second place, with 11 fungal species reacting positively by forming a yellow
halo around the fungal colony due to the degradation of complex carbohydrates
into simple sugars.
The widest halo refers to the
ability of fungi to secret the enzyme. The rat secretion rates went from low,
as in Macrophomina sp., to high secretion, as in Cladosporium sp.
(Table 3, Fig.2). The results of statistical analysis showed significant
differences (P<0.01) between the tested fungi in their ability to
produce cellulase enzyme. A large number of microorganisms are involved in the
degradation of cellulose.
However, the cellulase enzyme
plays a significant role in the biological activity of fungi, and they are
still one of the most essential microorganisms in cellulose degradation. It has
an extracellular enzyme system that breaks down cellulose into glucose that
dissolves in water and can be used as a source of energy. So, it’s played an
essential role in decomposing plant waste, in which cellulose forms
approximately 94% 29.
This
result agrees with the study of 30, which
showed nineteen species of fungi to grow on a cellulose medium.
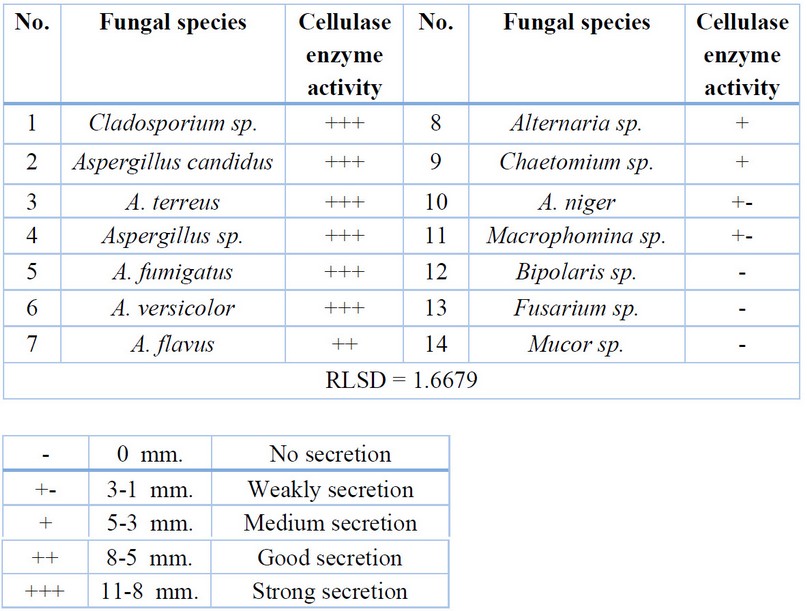
Table 3. Fungal enzymatic activity for cellulase enzyme
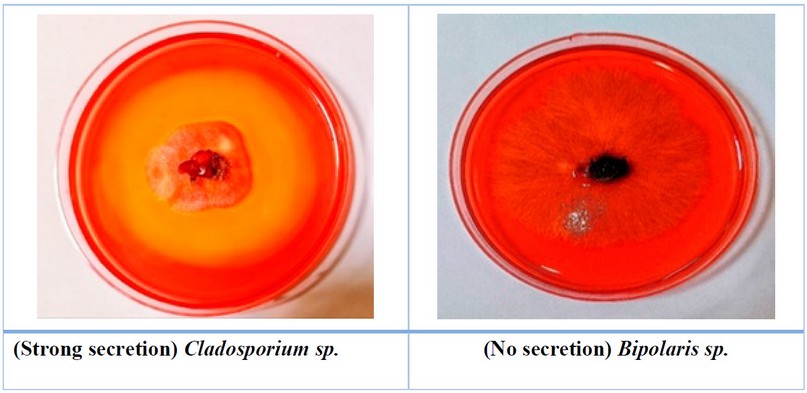
Figure 2. The ability of some
fungi to secret cellulase enzyme
The
lipase enzyme came in the third level, in which nine fungal species achieved
excellent efficiency in the secretion of this enzyme by forming a transparent
halo around the fungal colony due to the formation of a white precipitate or
white crystals.
The
ability to produce lipase enzyme appears with different capabilities of
secretion rates from low as in Chaetomium sp., to high secretion,
as in Cladosporium sp. (Table 4, Fig.3). The
results of statistical analysis showed significant differences (P<0.01)
between the tested fungi in their ability to produce lipase enzyme.
Several studies indicate the fungal ability isolated from
agricultural soil to produce lipase enzymes like 28, 32. And many fungi can
secrete this enzyme, and this may be because lipase is a fatty substance found
in grains and agricultural materials and can be used by fungi easily as a
nutritional source for growth, which contributed to the increase in the number
of fungi that were able to produce this enzyme, soils may also contain fatty
substances in the organic content of the soil, and microorganisms including
fungi, have an important role in the degradation of fatty substances through
the secretion of extracellular lipase enzyme
31.
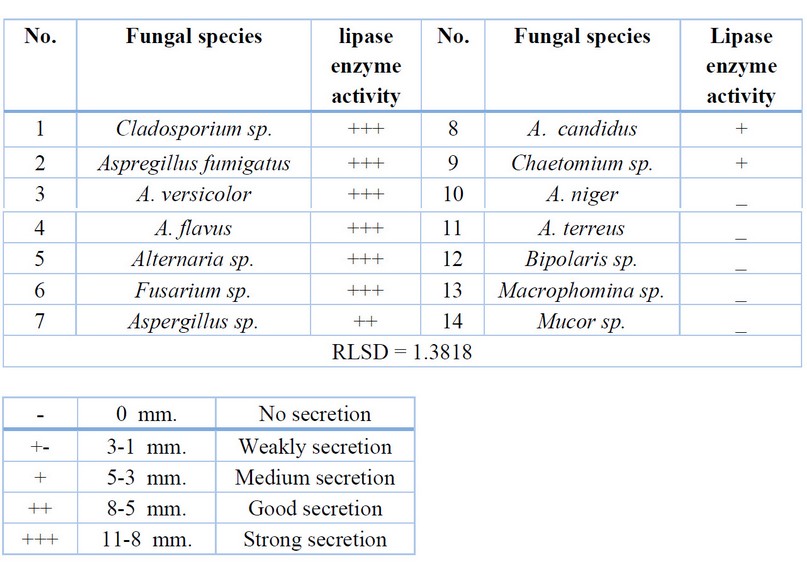
Table 4. Fungal enzymatic activity for lipase enzyme
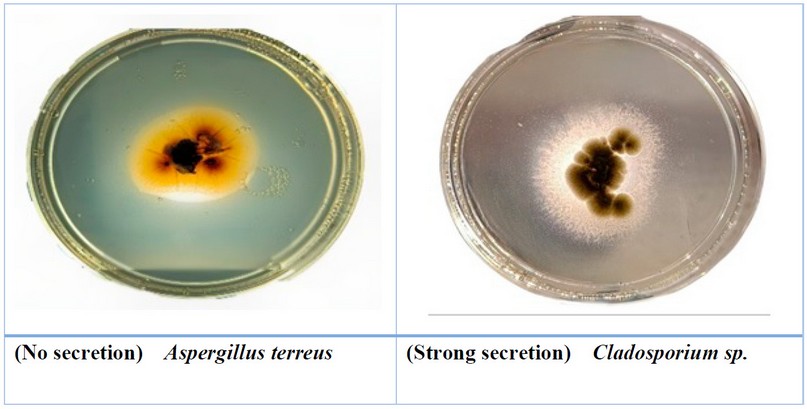
Figure 3. The ability of some fungi to
secret lipase enzyme
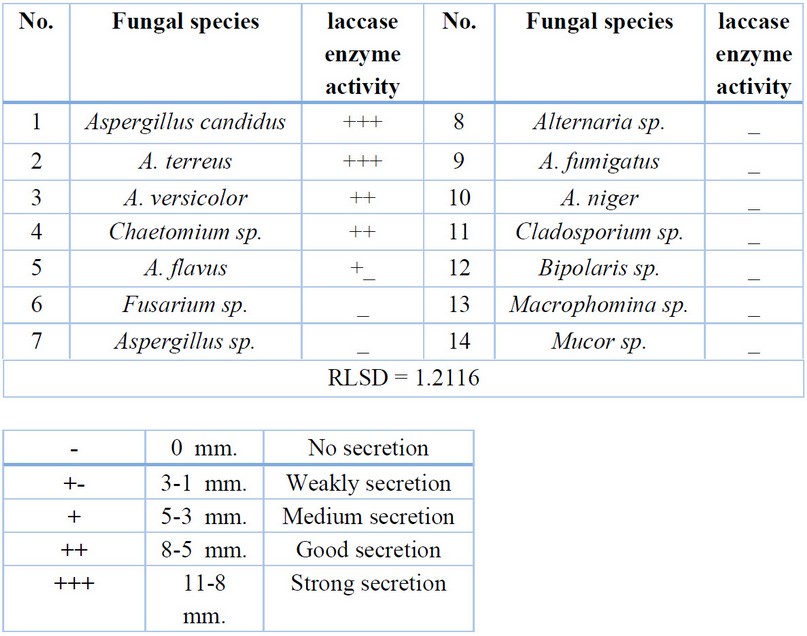
Table 5. Fungal enzymatic activity for laccase enzyme
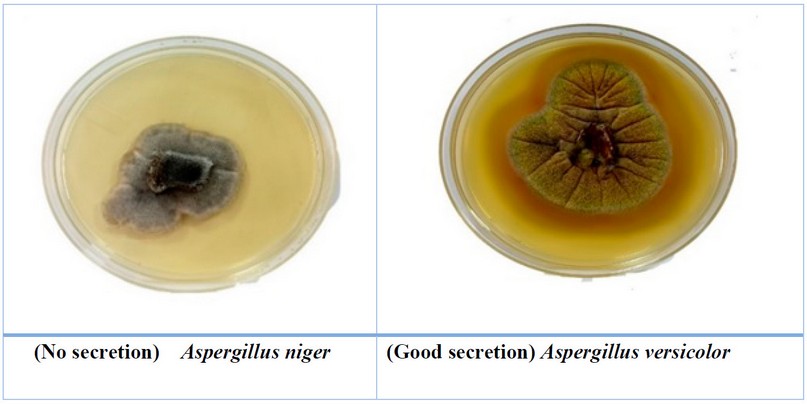
Figure 4. The
ability of some fungi to secret the Laccase enzyme
Five isolates only secrete the laccase
enzyme; Aspergillus candidus and A.
terreus fungi had the best secretion for this enzyme, while the other three
fungi produced it in very small or medium quantities Table 5, Fig. 4. The results of statistical analysis showed significant
differences (P<0.01) between the tested fungi in their ability to
produce laccase enzyme.
The
laccase enzyme plays a key role in the degradation of pollutants in the
environment due to the activity of free radicals during the oxidation of
aromatic compounds, phenolic compounds and amines, and this enzyme is used in
biotechnology applications as a biocatalyst and basidiomycetes are the best in
its secretion 33. In recent years, some studies have shown that some
anamorphic fungi could secrete this enzyme, which may be due to the mutations
resulting from the nature of the environment in which the fungi live 34.
This is consistent with the findings of 35, which found that very few fungi
can secrete this enzyme in his study. It also agreed with the study of 22,
which found only five species of fungi can secrete laccase enzyme.
CONCLUSIONS
In conclusion, the study found that the
ascomycetes fungi, particularly the genus Aspergillus, were the most isolated
in the agricultural soil samples. These fungi have high enzymatic activity and
the ability to secrete many reproductive units, allowing them to adapt well to
their environment and spread widely. Additionally, the study showed that the
tested fungi had varying abilities to secrete different types of enzymes, with
the manganese peroxidase enzyme being the most commonly secreted, followed by
cellulase, lipase, and laccase enzymes. These enzymes play important roles in the
environmental degradation of pollutants and organic materials.
Conflicts of
Interest: “The authors declare no
conflict of interest.”
REFERENCES
1.
Sunitha VH, Nirmala Devi
D, Srinivas C. Extracellular enzymatic activity of endophytic fungal strains
isolated from medicinal plants. World J. Agric. Sci. (2013); 9(1): 1-9.
2.
Pascual AR, Martín ME. Cellulose. Intech
Open. (2019); 22- 92.
3.
Sun
K, Li S, Si Y, Huang Q. Advances in laccase-triggered anabolism for
biotechnology applications. Crit. Rev. Biotechnol. (2021); 41(7): 969-993.
4.
Kavitha
K, Shankari K, Meenambiga S S. A review on extraction of lipase from
Aspergillus Species and its applications. RJPT. (2021); 14(8):4471-4475.
5.
Chowdhary P, Shukla G,
Raj G, Ferreira LFR, Bharagava R N. Microbial manganese peroxidase: a
ligninolytic enzyme and its ample opportunities in research. SN Appl. Sci. (2019);1: 1-12.
6.
Akhtar N, Mannan M A U.
Mycoremediation expunging environmental pollutants. Biotechnol. Rep.
(2020); 26: e00452.
7.
Benguenab
A, Chibani A. Biodegradation of petroleum hydrocarbons by filamentous fungi (Aspergillus
ustus and Purpureocillium lilacinum) isolated from used engine oil
contaminated soil. Acta Ecol. Sin. (2021); 41(5): 416-423.
8.
Wicklow D T, Whittingham W F. Soil microfungal
changes among the profiles of disturbed conifer‐hardwood forests. Ecol.
(1974); 55(1): 3-16.
9.
Raper
K, Fennell DI. The genus Aspergillus. Sec. ed. Robert Krieger Pybl. New
York. (1973): 686 pp.
10.
De
Hoog G S, Guarro J. Atlas of clinical fungi. CBS Netherland and university at
Rovira Virgili. Spain. (1995): 720pp.
11.
Sakagami Y, Watanabe R,
Aoyama C, Matsunaga S, Higaki N, Fujimura K. The intelligent ASIMO: System
overview and integration. IROS. September 2002; (3): 2478-2483.
12.
Sheifert K, Jones G M,
Games W, Kendrick B. The genera of hyphomycetes in Netherland. CBS-KNAW
fungal biodiversity center Utrecht. 2011; pp. 485.
13.
Guarro
J, Gene J, Stachigel AM, Figueras J. Atlas of soil Ascomycetes in Netherland.
CBS-KNAW fungal biodiversity center Utrecht. (2012); 997pp.
14.
Ireri N, Hamadi B I, Wanjiru W, Kachiru R.
Characterization, enzymatic activity and secondary metabolites of fungal
isolates from lake Sonachi in Kenya. J. Pharm. Biol. Sci. (2015); 0(2):65-76.
15.
Ali M I, Khalil N M,
El-Ghany M N A. Biodegradation of some polycyclic aromatic hydrocarbons by
Aspergillus terreus. Afr. J. Microbiol. Res. (2022); 6(16): 3783-3790.
16.
Laich F, Vaca I, Chavez
R. Rhodotorula portillonensis sp. nov., a basidiomycetous yeast isolated from
Antarctic shallow-water marine sediment. Int. J. Syst. Evol.
Microbiol. (2013); 63(Pt_10):
3884-3891.
17.
Al-Saadoon
A H, Al-Dossary M N. Fungi from submerged plant debris in aquatic habitats in
Iraq. Int. J. Biodivers. Conserv. (2014); 6(6):
468-487.
18.
Alrumman
S A, Standing D B, Paton G I. Effects of hydrocarbon contamination on soil
microbial community and enzyme activity J. King Saud Univ. Sci. (2015); 27(1): 31-41.
19.
Altaee M
S, Al-Dossary M A A. Evaluation of the enzymatic activity of some fungi
isolated from plastic contaminated soils and their LDPE biodegradation ability.
MRS Bull. (2021); 16(2).
20.
Taylor D L, Sinsabaugh R
L. The soil fungi: occurrence, phylogeny, and ecology. Soil microbiology, Biochem. Syst. Ecol. (2015);4: 77-109.
21.
Al-Saadoon
A H, Al-Dossary M A. Some fungi isolated from submerged plant debris in
southern Iraq. . MRS Bull. (2010);5(2): 207-221.
22.
Ali F T, Al-Dossary M A W. Study for the enzymatic
activity of some fungi isolated from agricultural soil. MRS Bull. (2019);14(1).
23.
Al-Dossary
M A W, Dahir A A. Bioethanol production from corn and barley wastes by Aspergillus
flavus. MRS Bull. (2023);18(1).
24.
Al-hamdani
R R, Al-Dossary M A. Investigation the Degradation Capabilities of Fungal
Isolate from Water and Sediment Samples to Congo Red Dye. Asian J. Environ. Sci.(2023);16(2).
25.
Patil M
G, Pagare J, Patil S N, Sidhu A K. Extracellular enzymatic activity of
endophytic fungi isolated from various medicinal plants. Int. J. Curr.
Microbiol. App. Sci. (2015); 4(3): 1035-1042.
26.
Jaiboon
K, Lertwattanasakul N, Limtong P,
Limtong S. Yeasts from peat in a tropical peat swamp forest in Thailand
and their ability to produce ethanol, indole-3-acetic acid and extracellular
enzymes. Mycol. Prog. (2016);15: 755-770.
27.
Usha K Y, Praveen K,
Reddy B R. Enhanced production of ligninolytic enzymes by a mushroom Stereum
ostrea. Biotechnol. Res. Int. 2014.
28.
Steudler S, Werner A,
Walther T. It is the mix that matters: Substrate-specific enzyme production
from filamentous fungi and bacteria through solid-state fermentation. Solid
State Fermentation: Int. j. Eng. Res. Appl. (2019); 51-81.
29.
Maruyama C R,
Bilesky-José N, de Lima R, Fraceto L F. Encapsulation of Trichoderma harzianum
preserves enzymatic activity and enhances the potential for biological control.
Front. Bioeng. Biotechnol. (2020); 8:
225.
30.
Jumaah E M, Al-Saadoon
A H, Al-Dossary M A. Enzymatic activity of some fungi isolated from submerged
plant parts in aquatic habitats southern Iraq. MRS Bull. (2020); 15(2).
31.
Bellaouchi R, Abouloifa
H, Rokni Y, Hasnaoui A, Ghabbour N, Hakkou A, Asehraou A. Characterization and
optimization of extracellular enzymes production by Aspergillus niger
strains isolated from date by-products. JGEB. (2021); 19(1): 1-8.
32.
Sopalun K, Iamtham S.
Isolation and screening of extracellular enzymatic activity of endophytic fungi
isolated from Thai orchids. S. Afr. J. Bot. (2020); 134: 273-279.
33.
Benitez S F, Sadañoski M
A, Velázquez J E, Zapata P D, Fonseca M I. Comparative study of single cultures
and a consortium of white rot fungi for polychlorinated biphenyls treatment. J. Appl. Microbiol. (2021); 131(4): 1775-1786.
34.
Han M,
Yang J, Ma J, Wang C, Chen S, Xu M, An Q. Extracellular laccase activity among
Ganoderma and Coriolopsis species grown on lignocellulosic wastes. Bioresour. Technol. (2022); 17(3):5049.
35.
Shah
H, Yusof F, Alam M Z. A new technique to estimate percentage decolorization of
synthetic dyes on solid media by extracellular laccase from white-rot fungus.
J. biorem. biodegrad. (2023); 27(1): 66-74.
Received:
26 September 2023 / Accepted: 15 April 2023 / Published:15 December 2023
Citation: Muhsen N A,
Al-Dossary M A. Diversity and
enzymatic activity of some fungi isolated from agricultural soil. Revis Bionatura 2023;8 (4) 31. http://dx.doi.org/10.21931/RB/2023.08.04.31
Publisher’s Note: Bionatura stays neutral concerning jurisdictional
claims in published maps and institutional affiliations.
Copyright: © 2023 by the authors. Submitted for possible
open-access publication under the terms and conditions of the Creative Commons
Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
