2023.08.03.78
Files > Volume 8 > Vol 8 No 3 2023
The clinical impact of chemokine receptor CCR5 Δ32 mutation in SARS-CoV-2 infected patients
1 Microbiology, College of Medicine, Al-Nahrain University, Baghdad, Iraq.
2 Microbiology, College of Medicine, Al-Nahrain University, Baghdad, Iraq.
3 Respiratory Medicine, Imamein Kadhimein, Medical City Baghdad, Iraq.
Correspondence: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.03.78
ABSTRACT
Since the first outbreak of coronavirus disease (COVID-19), many infected people have developed a severe infection, which is usually a sequel of cytokine overproduction. The chemokine receptor, such as chemokine receptor 5, also denoted as (CCR5) has a role in the pathogenicity of COVID-19 disease. The ongoing research paper tried to assess the impact role of CCR5Δ32 mutation in a group of Iraq SARS-CoV-2 infected patients. A total of 180 samples were enrolled in this study; 120 were patients infected with COVID-19 and verified by reverse transcriptase polymerase chain reaction (RT-PCR) in nasopharyngeal swabs. Those patients were categorized into two groups based on the severity of the disease: severe COVID-19, which included 60 patients and mild/moderate COVID-19 with 60 patients.
Furthermore, 60 subjects confirmed to be COVID-19-negative were enrolled in this study as a control group. The nucleic DNA was obtained from whole blood, and the CCR5Δ32 mutation was genotyped and detected by polymerase chain reaction using specific primer sequences. Results of the current study mentioned that out of the 180 samples in this study, 100 (100%) were wild type for the CCR5 gene (CCR5-wt), while none (0%) were mutant type for the CCR5-Δ32. This research has demonstrated that none of the study patients have the mutant CCR5 gene type (CCR5-32), assuming a lack of the role of CCR5Δ32 in the prognosis of COVID-19 infection.
Keywords: Chemokine receptor 5 (CCR5), CCR5Δ32 mutation, COVID-19, SARS-CoV-2.
INTRODUCTION
In March 2020, WHO declared COVID-19 a pandemic. The causal agent of this disease is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clinical manifestations range from asymptomatic to severe, critical cases and severe complications, which are highly frequent in elderly patients, especially those with comorbidity diseases such as (cardiovascular, obesity, diabetes mellitus, and renal diseases).1 Symptomatology of infection with SARS-CoV-2 could be determined by several viral factors, which include (viral load, mutation and virus linage), and host factors such as (immune system, sex, age, coinfection and nutritional status). 2
This disease reported high mortality rates worldwide with elevated levels of contagion through geographical areas, with different severity of infection, and the death rates from one region to another. Several factors cause this diversity, such as genetic factors of the virus, the genotype of human genes involved in the viral infection, and high levels of specific environmental pollutants.3
One of the crucial mechanisms associated with COVID-19 pathophysiology is the induction and recruitment of inflammatory immune cells towards infected lungs, which causes a subsequent release of different inflammatory mediators, including different cytokines such as IL-6, IL-1B, and TNF-α as well as chemokines which lead to a hyper-inflammatory state called cytokine storms .4 -6.
The chemokine receptor, such as chemokine receptor 5, also denoted as (CCR5) has a role in COVID-19 disease. It is an essential member of the G–protein–coupled receptor family abundantly present on the surface of different cells such as macrophages, monocytes, and T-cells. It has a role in the recruitment of leukocytes towards the site of inflammation and induction of inflammation in various infectious diseases. It also plays a role in viral infections such as Human immunodeficiency virus, Hepatitis B and C, West Nile Virus, and others. 7
The CCR5 gene is located on chromosome 3 at the short arm (p.21). A common 32bp deletion at the coding region causes the creation of a premature stop codon and produces a truncated protein with 215 amino acids length instead of 352 amino acids and significantly diminishes the surface expression of this receptor .8
The CCR5-D32 mutation was detected as a promising candidate to predict the COVID-19 severity. Different studies described the role of CCR5 gene polymorphisms that may impact susceptibility to COVID-19 infection.9 Similarly, another study shows a significantly higher frequency of CCR5Δ32 was observed in symptomatic COVID-19 patients compared to asymptomatic COVID-19 subjects. Moreover, these researchers found that this mutation may predict the SARS-CoV-2 severity.10 It is worth mentioning that the countries with higher mutation rates showed the highest viral infection and mortality rates .11, 12
This study strives to determine the impact of CCR5Δ32 mutation on susceptibility to COVID-19 among Iraqi patients.
MATERIAL AND METHODS
Study setting and population
The current investigation involved blood samples collected from two hospitals in Baghdad, which have frequently served as significant Quarantine centers in Baghdad and its suburbs, in addition to patients from other governors; those hospitals included Dar AL-Salam, Imamein Kadhimein Medical City; the samples collection in a period from (January 2021 to May 2021). Samples were processed in the Medical Microbiology College of Medicine AL- Nahrain University departments.
This case-control retrospective study was carried out during the 4th COVID-19 wave in 180 individuals with various age groups from both genders. Among the subjects in this study were 120 patients who presented signs and symptoms suggestive of having COVID-19 and tested positive for COVID-19, confirmed by reverse transcriptase polymerase chain reaction (RT-PCR) in nasopharyngeal swabs.
Patients' groups were categorized into two groups based on the severity of COVID-19 according to guidelines issued by the Iraqi Ministry of Health: a mild-moderate group composed of 60 patients and a severe group which included 60 patients. Other 60 healthy subjects confirmed to be COVID-19 negative by (RT-PCR) were recruited to represent a control group. Demographic variables, clinical characteristics and testing results for each patient's hematological and biochemical markers were obtained from the hospital registry.
Samples collection and handling
A blood sample equal to 5ml in a heparinized blood tube was collected from each member involved in this study and used for DNA Extraction by whole genomic extraction kit according to company instruction manual using PrestoTM mining DNA kit from Geneaid Company Taiwan and kept at (-20 Cº) until molecular assay.
Detected the CCR5Δ32 mutation
Quick Protocol SYNCTM DNA extraction Kit (Geneaid) has been applied to obtain highly pure genomic DNA using venous heparinized blood samples. The primer sequences (Forward and Reveres) as described by (Fath-Elrahman et al., 2022)13 F: (5’-CTG TGT TTG CGT CTC TCC CA) R: (5’-CCT CTT CTT CTC ATT TCG ACA). The reaction was completed in a 20 μl reaction that contained (1.5 μl) from each forward and reversed primer mixed with ready-to-use master mix, then added 3 μl from the DNA template; the volume was brought to 20μL by adding (14 μl) of nuclease-free water. PCR mixture without a DNA template (non-template negative control) was employed as a negative control. The thermal profile included 94°C one cycle for 3 min followed by 36 cycles of 95°C for 30 seconds, 62°C for 50 seconds, and 72°C for one minute .14 Five μl of PCR product was subjected to 1.5 % agarose gel electrophoresis with ethidium bromide (0.5μg /ml; Sigma). Amplicon product sizes were determined by comparison with a 100 bp DNA ladder (Bonier, Korea). Gene amplicon was detected at (222 bp) for the wild type and (190 bp) for the mutant type using an Ultra Violate after that photo was taken via a camera.
Statistical analysis
The data were examined using the social sciences statistical program (SPSS version 25, Chicago). Continuous data were subjected to a normality test (Shapiro-Wilk test). Measurements that followed a normal distribution were shown as the mean. Data that did not follow a normal distribution were shown as the median. The categorical variable was expressed as percentage and number and analyzed by chi-squared test, and the p-values less than 0.05 were considered statistically significant differences.
RESULTS
Demographic characteristics of the study population
Patients' mean ages with severe infection and mild/moderate infection were (60.90) and (38.92) years, respectively, which is a statistically significant difference compared to the mean of the healthy control group (30.50) years. While no statistically significant difference was found between the patients' groups and the control regarding gender (P=0.638).
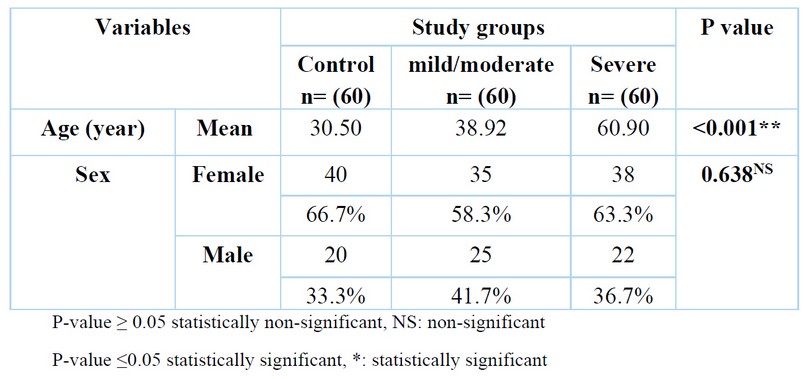
Table 1. The demographic characteristic of the study population
The chemokine receptor 5 mutation detection:
Out of the 180 samples assessed, which included 120 patients group and 60 healthy control group, 100 (100%) were wild type(222bp) for the CCR5 gene (CCR5-wt), while none (0%) was the mutant type(192bp) for the CCR5-Δ32 as shown in table (2) and figure (1).

Figure 1. The gel electrophoresis of CCR5 gene polymorphism (rs333) was amplified with a specific pair of primers using conventional PCR. Wild type on 222bp and mutant type on 192bp. L: ladder, NC: negative control, Lance (1-10): successful amplification with 222bp.
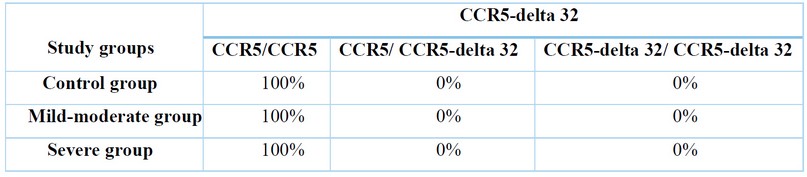
Table 2. Distribution of CCR5 genotype (heterozygous and homozygous) among COVID-19 patients' group and control group
DISCUSSION
Finding host genetic variables that promote vulnerability to or decrease resistance to complications of COVID-19 disease is the principal objective of this work. There are discrepancies between the findings of studies focusing on the role of the COVID-19 disease-causing CCR5delta 32 mutations and other studies focusing on CCR5WT. This gives the mutation a sense of justification. There is currently very little information directly relating to the impact of CCR5 polymorphisms on the severity of COVID-19. Furthermore, deterministic studies that offer logical explanations for how a mutation in the CCR5 gene would correlate with the severity and clinical manifestations of COVID-19 infection, including the recruitment of inflammatory and immune cells to the infection sites, are required. Moreover, according to our knowledge, no study in our country related to therapeutic management and support improved patient outcomes due to the finding of probable host genetic variation to COVID-19 infection.
This study was conducted with some limitations. For example, the difficulty of obtaining medical information for the most significant number of patients as a result of the medical quarantine conditions taken by most isolation hospitals, as well as the fact that this study included patients in a limited number of hospitals and the capital Baghdad only So, this does not represent the entire population of Iraq
COVID-19 is one of the crucial public health problems; it has ramifications for a large number of morbidity and mortality. The other studies reported that COVID-19 patients in advanced age were frequently associated with severe COVID-19 infection. In this study, the severity impact of COVID-19 patients was significantly higher in those older than 60. These findings agreed with previous studies from Iraq15, 16 and other countries in the world .17, 18 This finding may be explained byage–related disease comorbidities such as cardiovascular, diabetic and other chronic diseases. Moreover, pre-existing conditions such as smoking, obesity and other unhealthy lifestyles, which are most common in elderly patients, can exacerbate the risk of severity associated with age above 60 years19.
In the present study, there was not a considerable difference between severe and non-severe groups of COVID-19 patients in terms of gender. These findings agreed with previous studies from Iraq16. While inconsistent with the bulk of reports from Asia, where males were more likely to be afflicted by COVID-19 than females, with a male: female ratio of 1.1–1.93:1.20, 21 on the other hand distinguishably, in Europe, COVID-19- afflicted females more with a male: female ratio of 0.47:1 22, 23 the discrepancy in such results can attribute may be to differences in study sample size for each project.
Recent investigations have emphasized the significance of CCR5 in the etiology of COVID-19. Given that HIV-1 and SARS-CoV-2 share structural similarities, the anti-AIDS drug enfuvirtide has been suggested as a potential inhibitor of SARS-CoV-2 entry into cells .24, 25
In recent years, many studies have been done to highlight the importance of CCR5 and related mutations, particularly CCR5Δ32, in COVID-19 infection, if there are some protective effects of CCR5Δ32 against COVID-19 or its risk factor for COVID-19 infection severity. 26,27 It should be noted that there is a difference in the results of the researchers who dealt with this topic in their studies; some of them found that CCR5Δ32 mutation may have contributed to the severity of COVID-19 patients and explained this through Numerous leukocytes, including macrophages, T cells, and dendritic cells, express the chemokine receptor C-C chemokine receptor type 5 (CCR5), which has been demonstrated to be crucial in guiding leukocytes to the site of inflammation. Therefore, one of the critical checkpoints for enhancing inflammation in COVID-19 is CCR5; 11 or the role of CCR5 and ligand CCL5 interaction in the recruitment of the inflammatory immune cells to the site of infection and induction to produce the inflammatory cytokines particularly (IL-6, IL-1ß, IL-8, and RANTES) which causes cytokine storm .26
On the contrary, Hubacek JA et al. (2021) remarked that CCR5Δ32 is a protective factor for COVID-19 infection. They mentioned that the highest frequency of CCR5 32 carriers was found in SARS-CoV-2-positive/asymptomatic subjects (23.8%), followed by the lowest frequency in SARS-CoV-2-positive/symptomatic patients (16.7%), and the medium frequency in the control population (21.0%). So, they conclude that the CCR5 32 polymorphism could be able to predict how severe a SARS-CoV-2 infection will be .27
The current study reported that the CCR5 gene mutant type was not detected in all study groups (0%) enrolled in this project; on the other hand, the wild type (CCR5-wt) was seen in all study groups (100%), which indicates that CCR5Δ32 deletion has a significant negative correlation in the prognosis of COVID-19 infection. This study's observation of no CCR5Δ32mutation is consistent with a 1% earlier report by (Martinson et al., 1997) 28 and a more recent report by (Ekere et al., 2020) reported that; out of the 100 samples of HIV-infected subjects assessed, 100 (100%) were CCR5 wild type gene (CCR5-wt). 29
Also, the result is consistent with a European study by (Cizmarevic et al., 2021), who disclosed a negative association of the prevalence of CCR5-Δ32 with COVID-19 prevalence and mortality in the second European wave of the pandemic.12 Likewise, (Joel Henrique et al., 2020) declare that it is unlikely that CCR5Δ32 has some significant effect on the susceptibility or resistance to SARS-CoV-2 infection. However, CCR5 may have some impact on the clinical course of SARS-CoV-2 infection .30
From all existing data regarding the frequency and presence of CCR5Δ32 deletion, it is associated with race, and the rate of presence varies high frequency in Northern European countries. At the same time, East Asia and Africa have the lowest.31, 32 Such small percentages obtained in studies within the continent of Asia and Africa can explain why they were not found in our research, especially since the number of people participating in this study was 180. So, more significant numbers of COVID-19 cases and control groups are recommended to verify any impact of the CCR5 molecule on COVID-19 infection in our country.
CONCLUSION
This study concludes that the CCR5-Δ32 determines neither the risk of SARS-CoV-2 infection nor the course of the disease. Furthermore, This investigation disproved the theory that CCR5-32 played a role in the prognosis of COVID-19 infection by demonstrating the absence of the mutant CCR5 gene type in the individuals. While gender has little bearing on COVID-19 severity, age is among the crucial determinants.
Author Contributions
(1) Design and acquisition of data, Sample collected from patients and Statistical analysis.
(2) Writing the article and revising it critically for critical intellectual content analysis and interpretation of results.
(3) Patient selection and diagnosis
Funding
Self-funding
Institutional Review Board Statement
This project was granted ethical approval by the Iraqi Ministry of Health and Institutional Review Board (IRB) guideline at the College of Medicine of Al-Nahrain University.
Informed Consent Statement
Every patient engaged in this study gave their informed permission.
Data Availability Statement:
The datasets used and analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
The authors greatly appreciate the efforts of every employee at Al-Imammian Al-Kadhimein Medical City and the COVID-19 quarantine center in Baghdad during sample collection.
Conflicts of Interest
The author states that their interests do not conflict with one another.
REFERENCES
1- Hu, B.; Guo, H.; Zhou, P.; Shi, Z.L. Characteristics of SARSCoV-2 and COVID-19. Nat Rev Microbial 2021, 19,141–54.
2- Li, X.; Geng, M.; Peng, Y.; et al . Molecular immune pathogenesis and diagnosis of COVID-19.J. Pharm. Anal. 2020,10,102–108,
3- Mulder, C.; Conti, E.; Saccone, S.; Federico, C. Beyond virology: Environmental constraints of the first wave of COVID-19 cases in Italy. Environ. Sci. Pollut. Res. Int. 2021, 28, 31996–32004.
4- Jin, J.M.; Bai, P.; He, W.; et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020, 8, 152.
5- Qian, J.; Zhao, L.; Ye, R.Z.; Li, X.J.; Liu, Y.L. Age-dependent gender differences of COVID-19 in mainland China: comparative study. Clin Infect Dis, 2020, 71, 2488–2494
6- United Nations, Department of Economic and Social Affairs, Population Division. World Population Prospects 2019, Online edition. Rev. 1. 2019.
7- Klein, R.S. A moving target: the multiple roles of CCR5 in infectious diseases, J. Infect. Dis. 2008,197 183–186.
8- Mummidi, S.; Ahuja, S.S.; McDaniel, B.L.; Ahuja, S .K. The human CC chemokine receptor 5 (CCR5) gene. Multiple transcripts with 5'-end heterogeneity, dual promoter usage, and evidence for polymorphisms within the regulatory regions and noncoding exons. J. Biol. Chem. 1997, 272, 30662–30671.
9- Zang, R.; Gomez Castro, M.F.; McCune, B.T.; et al.TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020, 5, eabc3582.
10- Feng, S.; Song, F.; Guo, W.; Tan, J.; et al. Potential Genes Associated with COVID-19 and Comorbidity. Int. J. Med. Sci. 2022, 19, 402–415.
11- Panda, A.K.; Padhi, A.; Prusty, B.A.K. CCR5 D32 minor allele is associated with susceptibility to SARS-CoV-2 infection and death: An epidemiological investigation. Clin. Chim. Acta 2020, 510, 60–61.
12- Cizmarevic, S.N.; Tota, M.; Risti'c, S. Does the CCR5-D32 mutation explain the variable coronavirus-2019 pandemic statistics in Europe? Croat. Med. J. 2020, 61, 525–526.
13- Fath-Elrahman, M. H.; Alkarsany, M.; Nour, B .Y.; et al. Rating of CCR5-Delta 32 Homozygous Mutation in Sudanese HIV Patients and Sex Workers. World Journal of AIDS .2022, 12, 55-64.
14- Verma, R.; Gupta, R. B.; Singh, K.; et al. Distribution of CCR5Δ32, CCR2-64I and SDF1-3′A and plasma levels of SDF-1 in HIV-1 seronegative North Indians. Journal of Clinical Virology. 2007,38,198-203
15- Al-Hatemy, M.; Mohsin, M.; Al-Roubaey, D. The correlation between Interleukin-6 and D-dimer, Serum ferritin, CRP in COVID-19 patients in Al-Najaf province. Kufa Journal for Nursing Sciences .2022,12.
16- Jassim, S.I.; AL-Hasnawi, S. M .J.; Al-khaya, D. H. J. Decreased serum levels of chemokine receptors 1 & 5 in sever-critical Iraqi COVID-19 patients. Biochem. Cell. Arch. .2022, 22, 2545-2552.
17- Statsenko, Y.; Al Zahmi, F.; Habuza, T.; et al. Impact of Age and Sex on COVID-19 Severity Assessed from Radiologic and Clinical Findings. Frontiers in Cellular and Infection Microbiology. 2022, 11.
18- Du, R.H.; Liang, L.R.; Yang, C.Q.; et al. Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: a prospective cohort study. Eur Respir J. 2020,55, 2000524.
19- Lauc, G.; Sinclair, D. Biomarkers of biological age as predictors of COVID-19 disease severity. Aging. 2020, 12, 6490–1.
20- Meng, Y.; Wu, P.; Lu,W.; Liu, K.; Ma, K.; Huang, L. Sex-specific clinical characteristics and prognosis of coronavirus disease-19 infection in Wuhan, China: a retrospective study of 168 severe patients. PLoS Pathog. 2020,16.
21- Nikpouraghdam, M.; Farahani, A. J.; Alishiri, G.; Heydari, S.; Ebrahimnia, M.; Samadinia, H. Epidemiological characteristics of coronavirus disease 2019 (COVID-19) patients in IRAN: a single center study. J Clin Virol. 2020, 127,104378.
22- Lechien, J.R.; Chiesa-Estomba, C.M.; Place, S.; Van Laethem, Y.; Cabaraux, P.; Mat, Q. COVID-19 Task Force of YO-IFOS. Clinical and epidemiological characteristics of 1420 European patients with mild-to-moderate coronavirus disease 2019. J Intern Med. 2020 , 288,335–344.
23- Jia,J.; Hu, X.; Yang,F.; Song, X.; Dong, L.; Zhang, J. Epidemiological characteristics on the clustering nature of COVID-19 in Qingdao City, 2020: a descriptive analysis. Disaster Med Publ Health Prep. 2020, 14,1–5.
24- Ray, P.R.; Wangzhou, A.; Ghneim, N.; Yousuf, M.S.; et al. pharmacological interactome between COVID-19 patient samples and human sensory neurons reveals potential drivers of neurogenic pulmonary dysfunction. Brain, behavior, and immunity. 2020, 89,559-68.
25- Kliger, Y.; Levanon, E.Y. Cloaked similarity between HIV-1 and SARS-CoV suggests an anti-SARS strategy. BMC microbiology. 2003, 3,1-7.
26- Patterson, B.K.; Seethamraju, H.; Dhody, K.; et al. Disruption of the CCL5/ RANTES-CCR5 pathway restores immune homeostasis and reduces plasma viral load in critical COVID-19. medRxiv. 2020 doi.org/10.1101/2020.05.02.20084673. preprint. (Posted May 05, 2020; Revision under review).
27- Hubacek ,J.A.; Dusek, L.; Majek, O.; et al. CCR5Δ32 deletion as a protective factor in Czech first-wave COVID-19 subjects. Physiological research. 2021, 70,111.
28- Martinson, J.J.; Chapman, N.H.; Rees, D.C.; et al. Global distribution of the CCR5 gene 32-basepair deletion. Nature genetics. 1997, 16,100-3.
29- Ekere, E.F.; Useh, M.F.; Okoroiwu, H.U.; Mirabeau, T.Y. Cysteine-cysteine chemokine receptor 5 (CCR5) profile of HIV-infected subjects attending University of Calabar Teaching Hospital, Calabar, Southern Nigeria. BMC Infectious Diseases. 2020, 20,1-9.
30- Ellwanger, J.H.; Kulmann-Leal, B.; de Lima Kaminski, V.; et al. Beyond HIV infection: neglected and varied impacts of CCR5 and CCR5Δ32 on viral diseases. Virus research. 2020,286,198040
31- Kayeyi, N.; Sandøy, I.F.; Fylkesnes, K. Effects of Neighbourhood-Level Educational Attainment on HIV Prevalence among Young Women in Zambia. BMC Public Health. 2009, 9,310.
32- Tajbakhsh, A.; Fazeli, M.; Rezaee, M.; et al. Prevalence of CCR5delta32 in Northeastern Iran. BMC Medical Genetics .2019, 20: 184. DOI: 10.1186/s12881-019-0913-9
Received: 25 June 2023/ Accepted: 26 August 2023 / Published:15 September 2023
Citation: AL-Aziz Yousif Z A, Hassan J S, Hameed G H. The clinical impact of chemokine receptor CCR5 Δ32 mutation in SARS-CoV-2 infected patients .Revis Bionatura 2023;8 (3) 78. http://dx.doi.org/10.21931/RB/2023.08.03.78.
