2023.08.01.58
Files > Volume 8 > Vol 8 No 1 2023
Purification and characterization of lipase produced from Bacillus cereus (PCSIR NL-37)
Haniya Mazhar 1,2, #, Ali Afzal3, 
 , Syeda Eisha Hamid 3, Saira Ishaq6, Syed Shahid Ali 7, Hongxin Zhu 1,2, Zahid Hussain 8
, Syeda Eisha Hamid 3, Saira Ishaq6, Syed Shahid Ali 7, Hongxin Zhu 1,2, Zahid Hussain 8
1Bio-X-Renji Hospital Research Center, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, 160 Pujian Road, Shanghai 200127, China.
2Bio-X Institutes, Key Laboratory for the Genetics of Developmental and Neuropsychiatric Disorders, Ministry of Education, Shanghai Jiao Tong University, Shanghai, China.
3Molecular Medicine and Cancer Therapeutics Lab, Department of Zoology, Faculty of Sciences, University of Central Punjab, Lahore, Pakistan.
4University of Veterinary and Animal Sciences (UVAS), Lahore, Pakistan.
5Applied Molecular Biology & Biomedicine Lab, Department of Zoology, University of Narowal, Narowal, Pakistan.
6Department of Chemistry, Government College University, Lahore, Pakistan.
7 Institute of Zoology, University of Punjab, Quaid-e-Azam Campus, Lahore-Pakistan.
8 Institute of Industrial Biotechnology, Government College University, Lahore-54000, Pakistan.
*Corresponding Author: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.01.58
ABSTRACT
There is a growing trend to produce lipase from microorganisms owing to their commercial demand in various industries. Bacillus cereus has been shown to have extracellular lipase activity and high growth rates. This study explains the purification of microbial lipase to homogeneity by dialysis, precipitation and chromatography. The purified enzyme with 56kDa relative molecular mass exhibited the highest activity at 60°C (95.56U/ml) and pH 7 (124.50U/ml). The enzyme activity was highly promoted in the presence of K+ (136.17U/ml) and Zn++(133.07 U/ml), and SDS did not affect the enzyme activity, whereas in the company of triton X100 activity of lipase is maximum (23.90 U/ml). The enzyme activity was enhanced by using almond oil (120.00 U/ml) as a substrate. We deduce cheaper protocols for producing extracellular lipase via simple laboratory techniques, which could be a good insight for its production at the commercial level.
Keywords: Lipase; Bacillus cereus; chromatography; enzyme activity; purification.
EC 3.1.1.3 triacyl glycerol acyl hydrolase is a water-soluble enzyme that breaks down triacylglycerol into fatty acids and glycerol; the reactions are the oil-water interface.1 They have a proficient role in aminolysis, transesterification and esterification in organic solvents.2 The significant sources of obtaining lipase are microbes, plants and animals.
Microbial lipase – obtained from microorganisms, is produced by the fermentation of different bacterial species. Vakhlu and Kour (2006) mentioned that microbial lipases are much more stable than the lipases obtained from animals or plants.3 There are many advantages of microbial lipases as they have a low cost of production, easy to obtain and reliable in using biotechnological reactions. Many microbial species like Pseudomonas spp. Staphylococcus spp., Chromobacterium spp., Alcaligenes spp., Achramobacter spp. and Bacillus are widely used for lipase production at the commercial level.4
Following the production, the purification of lipase is of particular importance at the commercial level.5 The enantio specific and chemo regio behavior of lipases promote the researcher's and industrialists' interest in lipase studies. Lipases obtained from different sources have already been purified up to homogeneity. Several purifying strategies for fungal lipases are under investigation.6 Kumar et al. (2005) proposed DEAE-Sepharose column chromatography (diethyl aminoethyl-Sepharose column chromatography) and ammonium sulfate precipitation strategies for the purification (40-fold) of lipase from Bacillus coagulants.7 Usually employed ion exchangers are the carboxymethyl (CM) in cation exchange (20%) and the DEAE group in anion exchange (58%). Strong ion exchangers based on Q-Sepharose and triethyl aminoethyl groups are frequently used for protein purification.
Microbial lipases are essential as they can remove lipid stains applied in eco-friendly waste-water treatment 8; cold-adopted lipases are crucial.9 Moreover, personal care products utilize lipases to produce aroma compounds in cosmetic industries. Holding onto such demand, the present study is designed for a cost-effective lipase production with a significant focus on its purification and characterization.
Isolation and Screening of Lipolytic bacteria
Soil and water samples were collected from various sites, including oil industries, automobile garages and household wastages in Lahore, Pakistan. Samples were labeled as samples 1, 2 and 3 and stored at 4°C in sterilized air-tight containers. Lipase-producing strains from the samples were isolated using the serial dilution technique, and then screening was done by a tributyrin agar base medium.
Physical and biochemical characterization of isolates
Physical and biochemical characterization of isolates was done by Gram staining, Colony morphology, Urease test, Oxidase test, Catalase test, Tyrosine decomposition, Coagulase test, Starch hydrolysis, Sporulation test, Methyl red test, Citrate utilization, Nitrate reduction, Voges-Proskauer (VP) test, Indole test, Motility test, Growth in different temperatures and pH values and Acid release from sugars method.
Media for Lipase Production
The media selected for lipase production was composed of 5gm/l glucose, 5gm/l peptone, 5gm/l yeast extract, 5% Olive oil, 0.5gm/l MgSO47H2O, 3gm/l NaCl and 100 ml distilled water in a 250ml flask. This media was autoclaved at 120ºC for a half hour at 15 lbs. pressure and fermented at 37°C for 48 hours as a submerged fermentation method.
Lipase activity assay
Lipase activity was assayed using olive oil by the standard titrimetric method.10 Gum Arabic (5%w/v) in 5 ml of phosphate buffer of neutral pH and 2.0 ml of 0.6% CaCl2 solution were used for olive oil (10%v/v) emulsification while adding 500μl aliquot during the emulsion. The substrate mixture was incubated at 150 rpm and 30°C for 15 minutes in the rotary incubator. The reaction was terminated after incubation using ethanol solution: 3ml of acetone (1:1) and fatty acids were withdrawn. Phenolphthalein indicator was used during titration of fatty acids with 0.1NaOH at 10.5 pH for estimation of fatty acid liberation. One unit of enzyme is the quantity of enzyme required for hydrolysis of 1μmol of fatty acids from triglycerides.
Bradford Total Protein Assay
Bradford's (1976) 11 total protein assay was used to determine total protein concentration, a spectroscopic analytical way of determining protein concentration in a given sample.
Characterization of crude enzyme
Crude enzymes assayed the culture broth at optimum conditions of 30–70°C temperature for 10 minutes at a pH of 8.0. The residual enzyme activity was measured by using Standard enzyme test conditions.
Effect of pH on the activity and stability of lipase
Lipase activity was studied at 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, 8.0, 8.5 and 9.0 pH values using the following buffer solutions of 0.1M citrate phosphate buffer with pH, (3.5 - 6.0); 0.1 M sodium phosphate buffer with pH, (6.5 - 7.5); 0.1M tris HCl buffer with pH, (8.0 - 8.5); 0.1 M carbonate bicarbonate buffer with pH 9.0 with olive oil as substrate. The effect of pH on lipase activity was checked by incubating in desired buffers for 20, 40, 50, 60, 70, 80 and 90 minutes, respectively. 100μL aliquots of the buffered enzymes were used for assaying the lipase activity after incubation.
Effect of temperature on the activity and stability of lipase
Similarly, temperature impact was characterized at different ranges starting from 20°C to 80°C with a difference of 10°C each in olive oil. The lipase activity was assayed after incubating at the temperature mentioned above ranges for 30 minutes each.
Effects of different ions & detergents on lipase activity
According to previous studies, 1mM of some metal ions can affect enzyme activity. For studying this effect, different metal ions were also used. These ions include Zn2+, Ag2+, Cu2+, Fe2+, Na+, K+, Mn2+, Ni+, Co2+, Ca2+and Mg2+. The effect of these metals on enzyme activity was monitored by putting 1mM and 10mM of these metal ions in samples. Samples were stored for 1 hour at 30C. Similarly, the process has been performed in the case of chemical detergents. The enzyme activity was monitored after keeping the sample for 1 hour at 30C in the presence of detergents (DMSO, SDS, Tween 80 and trinton X-100).
Determination of substrate specificities
The effect of triacylglycerol on the activity of crude lipase was measured at 7.0 pH at 40°C. Different oils such as castor, almond, sunflower, and coconut oil were used as a substrate. As mentioned by Abdelkafi et al. (2009), emulsification of triolein was made just before use in 10 gum Arabic solutions.12
Purification of lipase enzyme Crude lipase enzyme preparation
The crude lipase enzyme was obtained from the culture broth of Bacillus cereus after its centrifugation at 10,000 rpm for 15 minutes. Lipase assay and protein estimation was done by using set methods.
Ammonium sulfate precipitation
Ammonium sulfate in solid form was added to the supernatant at the saturation of 20% and incubated at 25°C for 4 hours to precipitate ammonium sulfate on centrifugation. The same step was repeated at 40%, 60% and 80% saturation in the supernatant. The sediments were dialyzed by adding a small amount of de-ionized water repeatedly after regular intervals overnight. All of the concentrated fractions were tested for protein and enzyme activity to determine the fraction with the highest activity.13
Dialysis
After the precipitation, the buffer became loaded with much greater protein than before, bounded by the residual ammonium sulfate. The extra salts were dialyzed against a buffer in low salt concentration. First, the protein solution was put into a dialysis bag through small holes, and the protein remained inside the bag after the salt dialysis. The bag was placed on a large buffer and stirred for 16 to 24 hours, equilibrating the salt concentration. After repeating the equilibrium process several times, the salt concentration level in the bag will reach the minimum level. The same buffer was used to dialyze the precipitated solution for a night at 4oC by using a dialysis membrane -50 with a diameter of 14.3 mm, flat width of 24.26 mm, and capacity of 1 mL/cm. This dialyzed sample was used to assay lipase and protein estimation.
Column Chromatography
The dialyzed solution was put together on a cellulose powder column (2.5 × 15 cm) which was equilibrated with 10 mM phosphate buffer at neutral pH. After washing the column, the samples were eluted with a linear NaCl gradient (2 L), from 0 to 0.8M, in the same buffer with the flow rate adjustment to 0.5 mL/min (fraction volume: 5 mL). Lipase assay and protein estimation was done earlier. Finally, the lipase-rich fractions were pooled, stored at 4°C, and used as the purified lipase.
Determination of molecular weight by SDS PAGE
The molecular mass of purified lipase was determined by Sodium Dodecyl Sulphate Poly Acrylamide Gel Electrophoresis (SDS PAGE).14
After purification, SDS-PAGE was applied to samples as a separation process using a Thermo Scientific Electrophoresis device. 12% polyacrylamide gel prepared with 1.5 M Tris-HCl buffer, 30% acrylamide mixture, 10% (w/v) ammonium persulfate and TEMED and pH 8.8, 10% SDS (w/v). For sample loading, glass plates of size 10x10 were used, which had ten holes. The resolving gel was 12% polyacrylamide gel which constituted 2 ml of 30 % acrylamide mixture, 1.68 ml of water, 50 μl of 10 % SDS, 25 μl of 10 % ammonium persulfate, 1.25 ml of 1.5 M Tris-HCl (pH 8.8) and lastly 2.5 μl of TEMED up to 5ml of the final volume. Polymerization started after adding TEMED to the mixture and thus waited for another 10 minutes. The gel was dipped into distilled water to avoid dryness. For stacking gels, 4% gel consisted of 665μl of 30% acrylamide mixture in 3.05 ml of water, 50 μl of 10% SDS, 1.25 μl of 0.5 M Tris-HCl (pH 6.8), 25 μl of 10 % ammonium persulfate and lastly 5 μl of TEMED up to 5ml of the final volume. After polymerization, the ladder was removed from the top. The prepared gel was placed in a buffer tank filled with 1X Tris-Glycine-SDS (TGS) as a running buffer. Sample buffer was used to dilute the samples with the ratio of 1:4 (v/v) and turned it into blue-colored samples. The samples were heated in the water bath for 20 minutes at 95°C. Electrophoresis ran at 65 V constantly for 30 minutes, then at 100 V for 2 hours until the blue dye reached the bottom of the gel. After the completion of electrophoresis, the gel was transferred into a shallow staining tank filled with Coomassie colloidal blue staining solution for 24 hours with slow shaking. After staining, the gel was treated with 0.1M Tris-phosphate as a neutralization buffer. 25 %, v/v, methanol solution was used as a destaining agent, and 20 %, w/v, of ammonium sulfate was used as a fixing agent. The photographs were taken after staining and destaining the gel.
RESULTS
Isolation and Screening of Lipolytic Bacterial Strains
Samples collected from different sources were examined following serial dilutions. 100 bacterial isolates were separated, as shown in Table 1. Lipase-producing strains were isolated based on lipolytic activity; 25 bacterial isolates had the potential for lipase production.
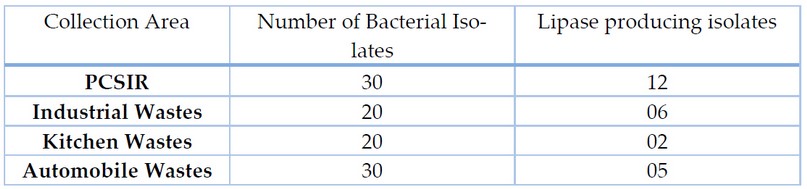
Table 1. Number of Bacterial Isolates from various sources
Screening and Identification of Lipase producing Isolates
Different qualitative and quantitative screening methods were applied to select the best lipase producers. Out of 100 suspected colonies, 25 bacterial isolates showed false lipase production and one with maximum production was selected for further studies. To the results of different qualitative and quantitative assays, the isolate NL-37 led total lipase production; the other strain did not exhibit considerable lipase activity (Table 2). Identification was carried out by morphological, biochemical and 16S rDNA sequence analysis (Table 3).
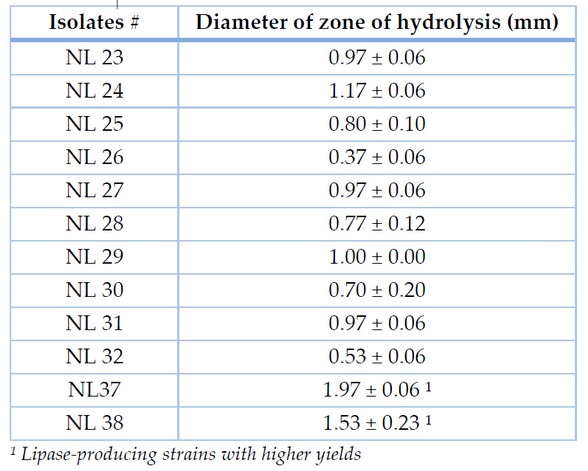
Table 2. Diameter of zones of hydrolysis by a crude enzyme on tributyrin agar plate from PCSIR after 24 hours
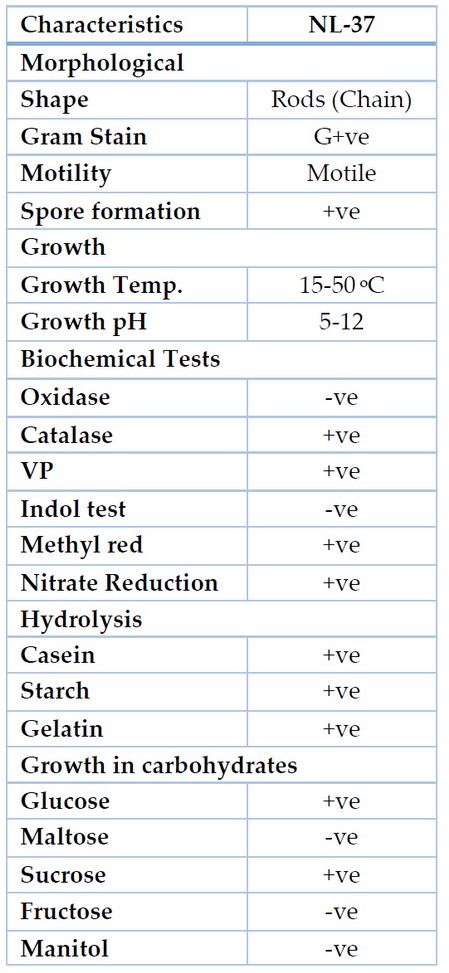
Table 3. Morphological and biochemical characterization of bacterial isolates
Purification of Extracellular Lipases
Ammonium Sulfate Precipitation
The enzyme produced after optimization of the cultural conditions was subjected to ammonium sulfate precipitation for salt-induced precipitation of proteins. 80% supernatant ammonium sulfate showed the enzyme activity of 23.3 U ml-1 by Bacillus cereus. After the desalting process, enzyme purification and recovery by Desalting is carried out to remove the traces of salt in sediments, resulting in increased enzyme activity.
Ion Exchange Chromatography
The desalted enzyme was subjected to a DEAE-cellulose column for ion exchange chromatography. The maximum lipase activity increased from the total fractions taken 13th fraction in the elution mechanism. At the same time, the molecular weight was determined by SDS PAGE, which showed 56 kDa of apparent weight.
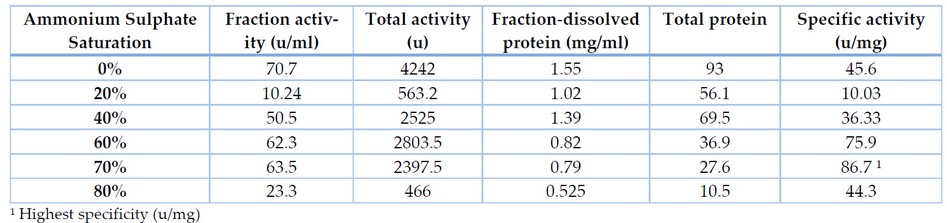
Table 4. Purification of lipase produced from Bacillus cereus at different concentrations of Ammonium Sulphate
The extracellular lipase of NL-37 was purified employing the ammonium sulfate precipitation method followed by dialysis and ion exchange chromatography. Table 4 shows that the supernatant's 70% ammonium sulfate saturation precipitated the highest amount of lipase. It offers 29.6% recovery yield with a 1.9 purification fold. Ammonium sulfate precipitation was followed by ion exchange chromatography. The purified lipase of NL-37 exhibited a final specific activity of 92 U/ml with 2.01-fold of purification and a recovery yield of 12.3% (Table 5). Low yield of the purified enzyme due to loss during ammonium sulfate precipitation.
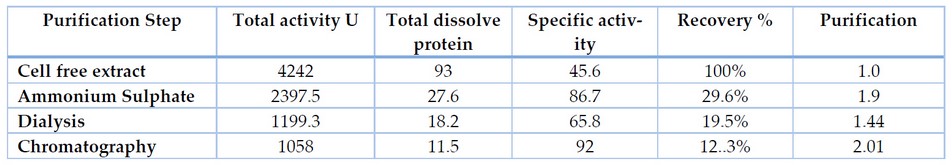
Table 5. Purification of extracellular lipase from Bacillus cereus
Characterization of Crude Lipases
pH Optimization of Extracellular Lipases
The effect of pH on the activity of crude lipases is shown in Figure 1a. The result of different pH values on the lipolytic activity of the enzyme was studied by incubating the enzyme in various buffers with a pH range of 4.0 to 10.0. After one hour of incubation, residual lipolytic activities of bacterial strains were measured using a titrimetric assay. It is evident from the results that the maximum residual activity by Bacillus cereus was 124.50% in the reaction mixture of pH 7.0 (Figure 1b).
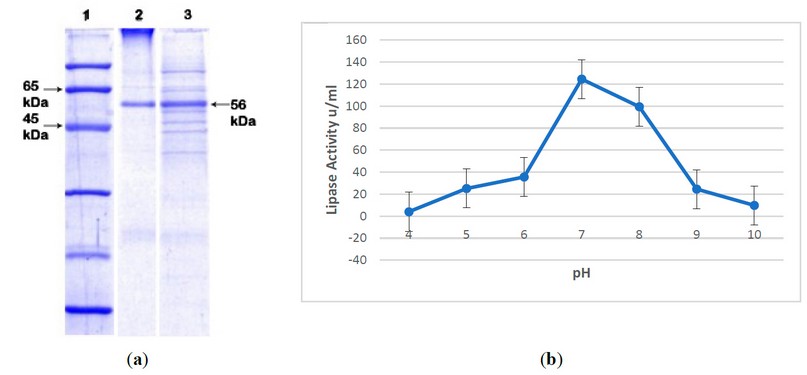
Figure 1. Characterization of Crude Lipase. (a) In pH optimization, SDS PAGE showed molecular weight: Line 1; molecular marker (fermentas), Line 2; purified Lipase, Line 3; lipase after ammonium sulfate precipitation; (b) Maximum lipase activity was shown at pH 7.0.
Temperature Optimization of Extracellular Lipases
In the present study, the enzyme extract was incubated at different temperature ranges, i.e., from 10-90°C (Figure 2). The effect of temperature on the activity of purified lipases was observed by incubating the enzyme at different temperatures for one hour. The results showed that lipases from Bacillus retained 90% of their activity at 60°C.
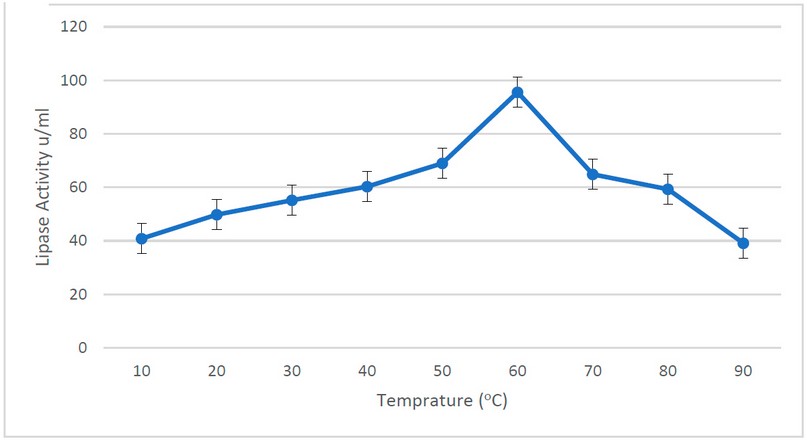
Figure 2. Lipase activity at different temperatures. The optimum temperature for lipase activity was 60°C, where the highest lipase activity was observed.
Effect of Metal Ions on the Activity
The residual activities were determined after incubation of the purified lipases with 1mM and 10mM metal ions (Figure 3a & 3b). At 1mM NL-37, lipase showed enhanced activity with Na+, Zn2+, Ca2+, K+ and Mg2+, but at a concentration of 10mM of metal ions, lipase activity of NL-37 decreased with Zn2+; however, increased with Ca2+, K+ and Mg2+.
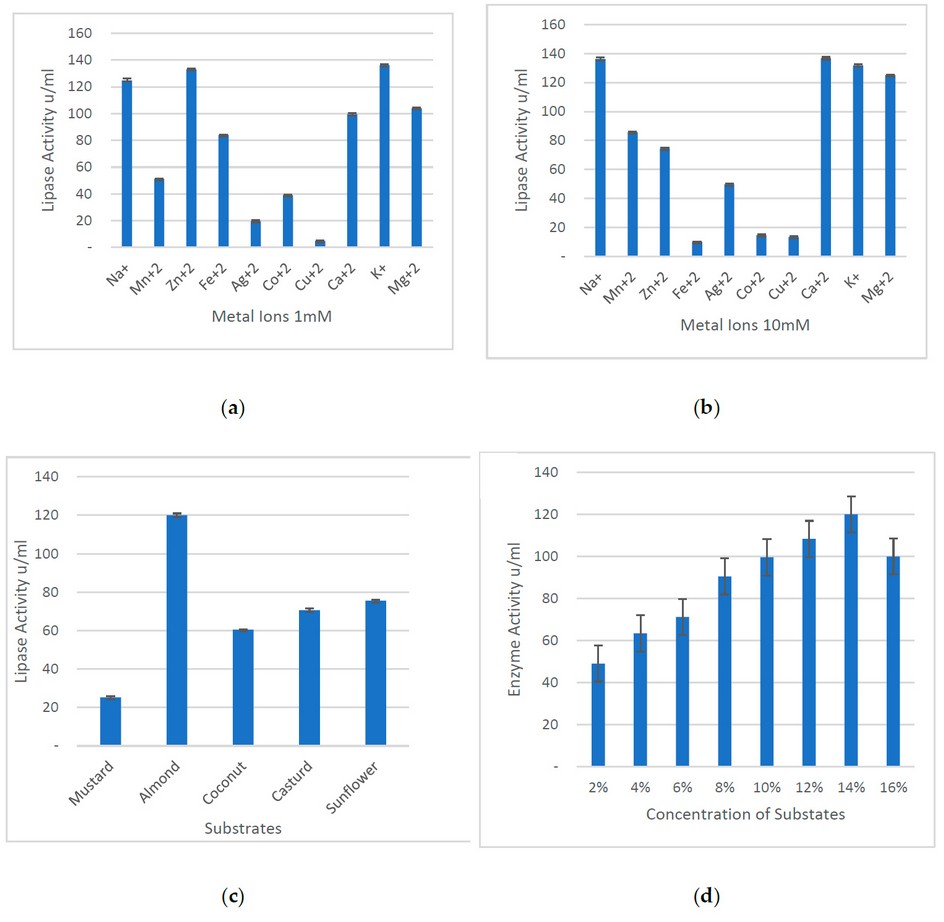
Figure 3. Characterization of lipase on various parameters. (a) At 1mM concentration of different metal ions, lipase showed the highest activity with Zn2+, K+, and Na+, whereas the least activity was with Cu2+ and Ag2+. On the other hand, (b) the lipase activity was higher in the presence of Na+, Ca2+, K+ and Mg2+. (c) Several substrates have various effects on lipase activity, as higher activity was shown with almonds, while the lowest activity was observed with mustard oil. (d) Specifically, olive oil showed higher lipase activity at a 14% concentration.
Effect of substrate on enzymatic activity
Different substrates were used to check the maximum activity of lipases. Other than olive oil, mustard, almond, sunflower, cast and coconut oils were used. All lipases obtained from Bacillus sp. showed maximum almond oil activity and less mustard oil (Figures 3c & 3d).
Our study utilizes different screening methods in lipase purification. Moreover, the previous reports 15 have indicated some critical steps, including the plate method using tributyrin and tween 80 with indicator. These methods are not reliable because the acidic waste metabolites alter the results. We tried to overcome this problem by adopting an alternate indicator called Rhodamine B.16 Bacillus spp. shows clear inflorescence in rhodamine B agar plates; for example, B. cereus has shown maximum lipase production at optimum conditions. In the present study, the lipase produced by Bacillus cereus had maximum activity at neutral pH. In support of these results, many scientists reported the neutral pH lipases 17, 18 or alkaline optimum pH. 19-22 Bacterial lipases are stable up to 60°C. In the case of present experiments, the lipases produced from Bacillus cereus showed maximum residual activity at 60oC, but lipase from the Bacillus strain did not completely denature even at 90°C. Although reports on bacterial lipases show a wide range of stability. In this study, almond oil as substrate showed a high percentage of activity, whereas very little activity was shown at mustard oil substrate specifically for Bacillus sp. 17, 23, 24
For the final application of lipase in pharmaceuticals, cosmetics, food and chemical industries certain degree of purity is required. In this observation, the lipases produced by Bacillus cereus were purified by ammonium sulfate precipitation; their specific activity was 200U/ml at 80% precipitation. Various strategies used to purify lipase have been reported 25-28 showing the optimal purification scheme for different microbial lipases.
The recovery of lipases was low due to loss during ammonium sulfate precipitation.29 After that, dialysis was performed and followed by DEAE-cellulose ion exchange chromatography. Results of chromatography showed a specific activity of 807.4 U/mg for Bacillus cereus. After purification, the SDS-PAGE analysis showed the molecular weight of purified protein obtained from Bacillus cereus was 56 kDa, while the purified form of the enzyme showed 40 kDa of apparent size in Octyl-Sepharose column chromatography.12
Lipases produced from Bacillus sp. are used as bio detergents and bioremediation of wastewater, and thus there is an augmenting demand for extracellular lipase production from microbial sources. In a nutshell, the present study utilizes standard laboratory procedures of purification, ensuring relatively higher specificity and recovery yields as well as characterization of microbial lipase at various parameters in a cost-effective manner, making it a promising insight for purification and characterization on an industrial level.
Supplementary Materials: Not applicable
Author Contributions: Conceptualization, Supervision and Intellectual Input, M. B. Khawar; Methodology and Writing-original draft, H. Mazhar, S. Aman and A. Afzal; Editing and Reviewing, M. B. Khawar and A. Afzal, S. E. Hamid, S. Ishaq, S.S. Ali, H. Zhu, and Z. Hussain.
Funding: This research received no external funding.
Institutional Review Board Statement: Not Applicable.
Informed Consent Statement: Not Applicable.
Acknowledgments: The authors are thankful to the Vice Chancellor of the University of Narowal for providing the platform for the accomplishment of this study.
Conflicts of Interest: The authors declare no conflict of interest.
1. Mehta, A., et al., The lipases and their applications with emphasis on food industry, in Microbial biotechnology in food and health. 2021, Elsevier. p. 143-164.
2. Chandra, P., R. Singh, and P.K. Arora, Microbial lipases and their industrial applications: a comprehensive review. Microbial Cell Factories, 2020. 19(1): p. 1-42.
3. Vakhlu, J., Yeast lipases: enzyme purification, biochemical properties and gene cloning. Electronic Journal of Biotechnology, 2006. 9(1): p. 0-0.
4. Chandra, P., et al., Microbial lipases and their industrial applications: a comprehensive review. Microbial Cell Factories, 2020. 19(1): p. 169.
5. Adetunji, A.I. and A.O. Olaniran, Production strategies and biotechnological relevance of microbial lipases: a review. Braz J Microbiol, 2021. 52(3): p. 1257-1269.
6. Alabdalall, A.H., et al., application and characterization of crude fungal lipases used to degrade fat and oil wastes. Scientific Reports, 2021. 11(1): p. 19670.
7. Kumar, S., et al., production, purification, and characterization of lipase from thermophilic and alkaliphilic Bacillus coagulans BTS-3. Protein expression and purification, 2005. 41(1): p. 38-44.
8. Shamim, S., U. Liaqat, and A. Rehman, Microbial lipases and their applications–a review. 2663-1040, 2018. 2(1).
9. Buchon, L., et al., Temperature dependence of extracellular enzymes production by psychrotrophic and psychrophilic bacteria. Biotechnology Letters, 2000. 22(19): p. 1577-1581.
10. Arzoglou, P., et al., Transferability of Lipase Titrimetric Assays: Deductions from an Interlaboratory Study. European Journal of Clinical Chemistry and Clinical Biochemistry, 1994. 32(10): p. 773-778.
11. Bradford, M.M., A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 1976. 72(1): p. 248-254.
12. Abdelkafi, S., et al., In vitro comparisons between Carica papaya and pancreatic lipases during test meal lipolysis: potential use of CPL in enzyme replacement therapy. Food Chemistry, 2009. 115(2): p. 488-494.
13. Massadeh, M., et al. Purification of lipase enzyme produced by Bacillus stearothermophilus HU1.
14. Laemmli, U.K., Cleavage of structural proteins during the assembly of the head of bacteriophage T4. nature, 1970. 227(5259): p. 680-685.
15. Beisson, F., et al., Methods for lipase detection and assay: a critical review. European Journal of Lipid Science and Technology, 2000. 102(2): p. 133-153.
16. Kouker, G. and K.-E. Jaeger, Specific and sensitive plate assay for bacterial lipases. Applied and environmental microbiology, 1987. 53(1): p. 211-213.
17. Dharmsthiti, S. and S. Luchai, Production, purification and characterization of thermophilic lipase from Bacillus sp. THL027. FEMS microbiology letters, 1999. 179(2): p. 241-246.
18. Lee, D.-W., et al., Isolation and characterization of a thermophilic lipase from Bacillus thermoleovorans ID-1. FEMS Microbiology Letters, 1999. 179(2): p. 393-400.
19. Schmidt-Dannert, C., M.L. Rúa, and R.D. Schmid, Two novel lipases from thermophile Bacillus thermocatenulatus: Screening, purification, cloning, overexpression, and properties, in Methods in enzymology. 1997, Elsevier. p. 194-220.
20. Sidhu, P., et al., production of extracellular alkaline lipase by a new thermophilicBacillus sp. Folia microbiologica, 1998. 43(1): p. 51-54.
21. Kanwar, L. and P. Goswami, Isolation of a Pseudomonas lipase produced in pure hydrocarbon substrate and its application in the synthesis of isoamyl acetate using membrane-immobilised lipase. Enzyme and Microbial Technology, 2002. 31(6): p. 727-735.
22. Sunna, A., et al., Biochemical characterization of a recombinant thermoalkalophilic lipase and assessment of its substrate enantioselectivity. Enzyme and microbial technology, 2002. 31(4): p. 472-476.
23. Sugihara, A., T. Tani, and Y. Tominaga, Purification and characterization of a novel thermostable lipase from Bacillus sp. The Journal of Biochemistry, 1991. 109(2): p. 211-216.
24. Lanser, A.C., L.K. Manthey, and C.T. Hou, Regioselectivity of new bacterial lipases determined by hydrolysis of triolein. Current Microbiology, 2002. 44(5): p. 336-340.
25. Antonian, E., Recent advances in the purification, characterization and structure determination of lipases. Lipids, 1988. 23(12): p. 1101-1106.
26. Taipa, M.A., M.R. Aires-Barros, and J.M.S. Cabral, Purification of lipases. Journal of biotechnology, 1992. 26(2-3): p. 111-142.
27. Cygler, M., J.D. Schrag, and F. Ergan, Advances in Structural Understanding of Lipases. Biotechnology and Genetic Engineering Reviews, 1992. 10(1): p. 143-184.
28. Yagmurov, E.R., G.V. Kozlov, and MA Pushkarev, Lipase purification: The review of conventional and novel methods. Journal of Hygienic Engineering and Design, 2017. 20: p. 60-69.
29. Romdhane, I.B.-B., et al., A novel thermoactive and alkaline lipase from Talaromyces thermophilus fungus for use in laundry detergents. Biochemical Engineering Journal, 2010. 53(1): p. 112-120.
Received: January 15, 2023 / Accepted: February 25, 2023 / Published:15 March 2023
Citation: Mazhar H, Afzal A, Aman S, Babar Khawar M, Eisha Hamid S, Ishaq S, Shahid Ali S, Zhu H, Hussain Z. Purification and characterization of lipase produced from Bacillus cereus (PCSIR NL-37). Revis Bionatura 2023;8 (1) 58. http://dx.doi.org/10.21931/RB/2023.08.01.58
