2022.07.01.2
Files > Volume 7 > Vol 7 No 1 2022
Indicators to achieve a better positioning of Ecuadorian scientific journals
1 Pontifica Universidad Católica del Ecuador Sede Ibarra, Imbabura, Ecuador.
* Corresponding Author [email protected];
Available from: http://dx.doi.org/10.21931/RB/2022.07.01.2
ABSTRACT
Ecuador's scientific journals are not in a good position worldwide, as seen in the small number of journals indexed in impact databases. This research was carried out due to the current and prevailing need to improve the performance level of national scientific journals at the international level. This work sought to generate the information necessary for Ecuadorian scientific journals to have a greater scope considering the criteria for indexing in the Scopus, Clarivate Web of Science, Scielo, and Redalyc databases. This was determined based on the hypothesis: Failure to comply with the indicators for international scientific databases makes Ecuadorian journals not part of these collections. We identified differences between public and private institutions and the status of communication journals in compliance with the same criteria. We analyzed 55 Ecuadorian scientific journals already indexed in Latindex. The analysis was carried out by creating matrices that made it possible to quantify the fulfillment of the criteria, implementing a categorization of Yes for the journals that meet the indicator or NO for those that do not. Similarly, interviews were conducted with editors of the Ecuadorian journals with the best indexing to triangulate the information discussed in this research. The research showed that, although most magazines meet the vast majority of indexing criteria, the main problems are the geographic and institutional diversity of the editors and authors in the journals.
Keywords. Databases, Indexing, Ecuadorian scientific journals, Scopus, Clarivate Web of Science, Scielo and Redalyc.
INTRODUCTION
Scientific journals are the basis of the knowledge community. They are in charge of collecting all the research that drives human beings as knowledge generators. These journals represent the vital means to disseminate research results and usually specialize in different disciplines or academic sub-disciplines1.
Despite the importance of scientific journals, one of the problems within the academic context is identifying journals with a high impact. The great variety of journals at different levels of relevance leads to which articles provide a more significant impact and, therefore, better positioning in the ranking2.
With the introduction of the 2008 Higher Education reform, scientific production in Ecuador experienced a turning point. From that date on, the national government required all higher education institutions to be directly linked and promote scientific and technological research3. The new requirements by said reform and the control of these, by the hand of the higher education agency, have led to an exponential growth in scientific production in the country since its creation4.
Despite this exponential growth in scientific publications in Ecuador, this positioning is inferior within Latin America, occupying in 2020 the 9th position in terms of publications and indexed scientific journals, with only 3 of the latter. However, the difference between the number of articles published by Ecuador (30775 publications) and Peru (36414 publications), the following country in the ranking, does not represent a significant difference, considering that the number of indexed Peruvian journals is 135.
As observed in the aspects pointed out, we conducted this research due to the current and prevailing need to improve the level of performance of Ecuadorian scientific journals. In this way, analyzing and evaluating the indicators that make a journal has a more significant impact will mean the creation of a basis for Ecuadorian scientific journals to have greater relevance in the future.
We considered all the information presented on the Scimago web page regarding the above. SCImago Journal Rank (acronym SJR) derived a positioning indicator for Latin American journals until 2020. The web analysis shows the status of Latin American journals indexed in the Scopus database, whose measurement reported the position of each of the countries related to scientific production during that year.
The first indicator analyzed was the number of indexed documents. Brazil was ranked with 1,145,853 documents, while in second and third place are Mexico with 387,111 and Argentina with 247,088. Below these positions in the top 10 indicators are Peru with 36 414 in 8th place and Ecuador in 9th place with 30 775 indexed documents. Uruguay closes the Top 10 with 23,887 documents6.
However, using the Scimago Journal Rank method and adding another indicator referring to the number of journals indexed in Scopus shows how the positions change. For example, Brazil remains in first place with 389 journals, while Colombia has 113 journals. However, Mexico and Chile are close behind with 111 and Chile with 109, occupying positions 3 and 4. At the same time, in positions 8 and 9 are Peru with 13 journals and Ecuador with 3 indexed journals6.
Therefore, the validity of the results of this research will provide relevant information so that scientific journals in Ecuador can consider obtaining a better positioning, both those that are starting and those that have a history of academic dissemination. This will allow national research to have a greater reach and the Ecuadorian academic community to achieve a more significant international impact.
MATERIALS AND METHODS
Type of research
A basic approach and an explanatory level and field research are presented. Its purpose is to diagnose the situation of scientific journals in Ecuador and determine what parameters should be worked on to improve the positioning and indexing of journals.
Besides having a documentary and field design, the research was developed with a synchronic strategy, based on the current context, since the objective of defining "why" Ecuadorian scientific journals do not have better positioning was a primary element in the whole research process.
The data collection was derived from the field observation technique, allowing us to understand two phases of the research. The first one, to observe and interact directly with the organizational achievement of the subject of study, allowed to determine how it behaves in its specific environment with the general objective. The second phase made it possible to identify how the different variables in this environment can interact with the subject.
It is important to emphasize that the work is also configured with aspects of documentary research. To obtain the necessary data, texts, documents, and other documentary elements were used to analyze and interpret them critically to generate new knowledge.
Methods
A mixed approach (quantitative and qualitative) was chosen using the analytical-synthetic research method. The objective is to analyze all the leading databases' indexing criteria, synthesize the results, and propose new approaches and strategies to better position in Ecuadorian popular science journals. For this purpose, descriptive research was carried out to specify properties, characteristics and important features that make a scientific journal good positioning.
The use of this method involved collecting data from all available digital sources. The data synthesis will be displayed employing tables, charts, graphs, or any non-numerical form to facilitate understanding of the data collected. The studied indicators were obtained from the following databases: Scopus, Clarivate, Latindex, Scielo and Redalyc.
Population and sample
The sample size is 55 indexed Ecuadorian scientific journals. The journals were selected using a series of criteria detailed below. First, the journals of the institutions present in the Scimago university ranking were selected since this objectively gives us the context of the universities according to their scientific production, taking into account 3 parameters, performance in terms of research, innovation and social impact (De Moya Anegón et al., 2020). To reduce sample size and add another parameter to analyze, the institutions of the provinces of Ecuador where there were public and private institutions within the Scimago ranking were selected. Following these criteria, we chose journals of institutions from Pichincha, Guayas and Azuay.
We obtained a final sample size of 55 journals distributed among 15 institutions following this selection criterion. Of these, 7 institutions are public and have 27 journals in the sample, and 8 private institutions have 28 journals. This homogeneous distribution among the sample allows a comparison between the 2 types of institutions.
In addition to the 3 provinces mentioned, another selection criterion was included, which stipulated considering the geo-referential locality of the Universidad Católica del Ecuador, Ibarra campus through the university's journal, indexed in Latindex. This parameter inclusion was because the present research is part of a final degree work for obtaining the academic degree.
Content analysis
The content analysis was carried out based on compliance with the indexing criteria of the Scopus, Clarivate, Scielo and Redalyc databases. To quantify compliance with the criteria, we implemented a binary categorization system, assigning Yes for journals that meet the indicator or No for those that do not. All these data were then quantified to create graphs that allow a better understanding of the situation of Ecuadorian journals. The journals analyzed correspond to the last volume published.
The information necessary to achieve indexing is publicly available and provided by each database. Of the parameters for indexing, we only chose those explicitly verifiable on each journal's web page. Since there are several criteria for which the intervention of teams of evaluators from these databases is required, these more complex parameters occur in the final indexing process. Meanwhile, the parameters analyzed in this research work are those the journals are in the first steps of the indexing process. If the journals did not comply with these, the analysis would have stopped, and it would not be necessary to consider the more complex ones.
Semi-structured interviews
We realized semi-structured interviews with the editors of the journals with the best indexing in Ecuador. These interviews support the information gathered in the content analysis since some indexing indicators cannot be appreciated as external agents of the indexing evaluation entities. Likewise, the editors provide details about the management of a journal, specifically all those related to achieving indexing, maintaining its status, and improving the journal's quality constantly to achieve a more significant impact.
The interviews were conducted with a guide of topics to be covered. To make them more informal and obtain as much information as possible, they were conducted in conversation mode, interweaving the information provided by the interviewee with the topics and questions of interest for the research.
We conducted the interviews once we finished the statistical analysis. With the results of this analysis, it was possible to establish a conversation with each of the editors with complete knowledge of the journal's situation. We also inquired about the unfulfilled indicators or how they met specific parameters that the rest of the journals could not achieve.
Matrix
Four matrix were created for each database since each has different indexing indicators. The following data were added to all the institutions in the sample: the province where they are located and the type of institution.
Interview schedule
A series of topics were determined to guide the conversation for the interviews. These were the following:
- Creation process
- Indexing problems
- Editorial committee
- Publication authors
- Number of articles and homogeneity
- Financing and support
- What the journal should improve
- Work between journals and faculties within the institution
- Other journals of the same institution
RESULTS
Indicator matrix
Fifty-five journals indexed in Latindex were analyzed to check compliance with indexing indicators in the Scopus, Web of Science, Redalyc and Scielo databases. The parameters were compiled in a matrix of indicators. According to whether or not they met these criteria, they were marked with YES or NO criteria to quantify the total number of journals that met these indexing guidelines for each database. A total of 4 tables were created, each one specific to each database (see tables in annexes). Most journals met the vast majority parameters, so their inclusion in the analysis process is not statistically relevant.
Outstanding indicators
Once we collected all of the necessary data, we analyzed indicators using tables and their respective graphs to evaluate the Ecuadorian journals in terms of their indexing according to the indicators that stood out the most. It was also reviewed for a complete understanding and a deeper analysis to see any difference between public and private institutions. In addition, as this research is a thesis of the social communication career, the current situation of the country's communication journals will be analyzed. We rounded all values to the next higher value for calculating percentages, considering two significant figures.
Authors and editors diversity
Publishers and authors' geographic diversity are the parameters that stand out most for their non-compliance in the Scopus and Clarivate databases. At the same time, the Redalyc and Scielo databases have similar indicators, but they do not focus on geographic diversity but institutional diversity (Figure 1).
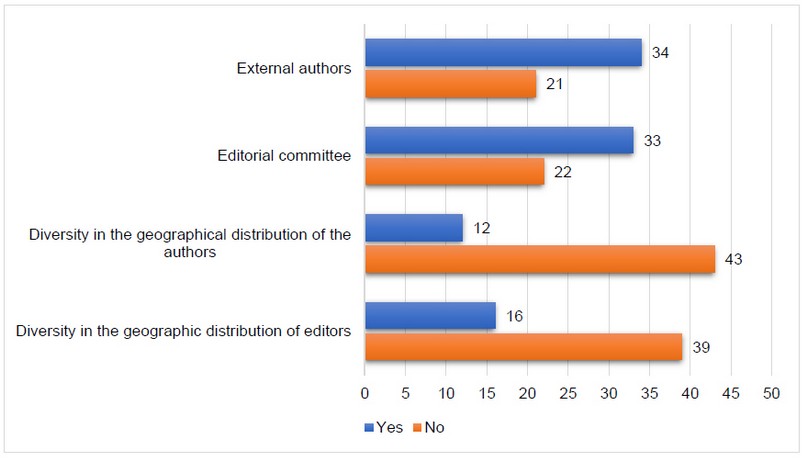
Figure 1. Authors and editors diversity
Information presentation on the website
Poor display of the journal's information on their websites was another indicator not fully met. Figure 2 shows the journals with web content in English and tables of contents and display formats.

Figure 2. Information presentation on the website.
Quantity and homogeneity of articles in the journal's issues
Other critical indicators to consider are the number of articles and their homogeneity to understand their impact on the journals' quality and scope (Figure 3).

Figure 3. Quantity and homogeneity of articles in the journal's issues
Availability and visibility of the information
Regarding the parameters of information's availability and visibility on their digital platform, some missing criteria in several journals could also be appreciated, such as: Their document's digital preservation policy, whether they are hosted on an independent electronic platform, the presence of statistical data and the distinction of current indexing records (Figure 4).

Figure 4. Availability and visibility of the information
Journal and author information
Finally, the journal and author information indicators have a significant percentage of non-compliances. These can be seen in Figure 5, which shows the use of bibliographic mastheads, author identifiers, and digital resource identifiers present in the journals.

Figure 5. Journal and author information
DISCUSSION
Authors and editors diversity
One of the main points to achieve the indexing of scientific journals according to the indexing guidelines of the 4 databases analyzed, Scopus, Clarivate, Scielo and Redalyc, is the diversity of the editorial team and the diversity of the authors who publish in the journal. The relevance of the editorial's team geographical diversity is reflected when promoting the research output of the journals. Therefore, journals with an editorial team of various regions attract greater geographic diversity7.
Both Scopus and Clarivate require a minimum of 70% geographic diversity, both the journal editorial team and the authors publishing in the journal. As shown in Figure 1, most of the journals analyzed do not meet these indicators. Only 16 out of the 55 journals meet the editor's geographic diversity criterion equivalent to approximately 29% of the sample analyzed. This shows that there is little foreign participation in the editorial boards of Ecuadorian journals. At the same time, only 12 of the 55 journals (approximately 22%) have geographic diversity of publishers, which reveals practically no participation of foreign authors in Ecuadorian journals. These percentages show a strong correlation between the editorial team's diversity and the authors' diversity of publications, as Demeter & Gaspar stated in their research in 2018.
The Redalyc and Scielo databases slightly change the requirement of this indicator, making it more local, and require that the journals have 70% of authors and editors from outside the publishing institution. As shown in Figure 1, concerning the previous indicators, there is an improvement; however, it continues to be a very negative factor for the journals, which prevents their indexing. As for the editors, 33 of the 55 journals (66%) comply with this requirement since they should only have less than 30% of their editorial board coming from the same institution. A similar situation arises with the author's distribution, where 34 out of 55 journals (approximately 62%) comply with the parameter of having less than 30% of their articles submitted by members of the same institution. This shows an improvement to the indicators of the geographical diversity of editors and authors. However, it is still a very high percentage of non-compliance requiring only the editorial board and publishing authors to be external to the publishing identity.
All the editors interviewed expressed that these indicators are the most important for indexing journals in impact databases. The editors of the journals La Granja and Bionatura (the only journals in the sample indexed in Scopus) emphasize that the creation of editorial committees is based on the journals' management of contacts since, in the beginning, these committees were formed by contacts close to the editors.
Regarding the criteria for the distribution of authors, the editors interviewed have a similar approach to that of the distribution of editorial committees. They affirm that the diversity of the editorial committees in conjunction with works such as community managers and the use of social networks manage to attract the attention of potential authors to publish in their journals. Likewise, calls for papers such as the call for papers help receive articles. However, they emphasize that the vast majority comes from contacts and the rising journal's reputation.
They are comparing private and public institutions with these criteria. Regarding the geographic diversity of the editors, the private journals meet this criterion, with a total of 10 out of 28 journals (approximately 36%). In comparison, the public journals meet with 6 out of 27 journals (approximately 22%). Analyzing this parameter from the diversity of distribution of editors, in terms of whether they are part of the journal's institution or not, private journals comply with the parameter in 18 out of 28 journals (approximately 64%). In comparison, public journals comply with it in 15 out of 27 journals (approximately 56%). The distribution of authors presents a similar panorama since private institutions have only 8 out of 28 journals (approximately 29%) that meet this criterion.
In contrast, public institutions have only 4 out of 27 journals (approximately 15%) that meet it. As for authors from the same country but outside the journal, both types of institutions comply with 17 out of 28 journals (approximately 61%) in the case of private journals and 17 out of 27 among public journals (approximately 63%). Our results showed no noticeable difference between the types of institutions; however, private institutions have higher compliance with these criteria.
Information presentation on the web page
In recent decades, access to research articles has changed from print to digital sources, from closed to open access. Despite these changes and broader access to research results, there are still accessibility barriers regarding the formats available to access information8. English as the universal language of science has allowed scientists to communicate ideas freely and access the world's scientific literature9. These two previously mentioned characteristics are essential points to better indexing scientific journals since they should provide more feasible accessibility to the research present in each. These indexing points are the use of English in tables of contents and on the journal's home page, the display formats and the use of tables of contents or summaries.
In the journals analyzed, it can be seen in Figure 2, how a total of 40 of the 55 journals (73%) comply with the translation of the table of contents, and 42 of the 55 journals (76%) offer the possibility of translating the home page into English. This shows that the vast majority of the journals understand the importance of using the English language to achieve a greater reach for their publications. During the interviews conducted, some of the editors interviewed expressed that English was one of the journals' problems in achieving indexing in the Scopus database, which became one of the main criteria to be improved.
At the same time, as shown in Figure 2, 33 of the 55 journals (60%) present a diversity of display formats, which means that they have their articles in formats other than PDF, which is currently the most popular. Of the journals analyzed, many used other formats in previous issues. However, for the analysis presented, only the last published issue of the journal was considered, and at the time of the study, only 33 journals had at least one other format, other than PDF.
The use of tables of contents or summary is met in 87% (48 out of 55 journals) of the journals, making it an indicator that is met in the vast majority of cases. According to Ansorge (2021)10, not presenting different display formats creates barriers for readers. For him, solutions exist; for example, OJS allows creating an HTML version of the article. Nevertheless, this requires some additional work from publishers and payment for the use of the platform, which is probably why many scientific journals prefer only to provide full-text articles in PDF format.
As for the comparison between private and public institutions, public universities have better compliance with all indicators except for the translation of the table of contents; however, it is a slight difference.
Quantity and homogeneity of articles in the journal's issues
For indexing, the databases analyzed require a minimum number of articles published per year by the journal and homogeneity in the number of articles published per year. Although the homogeneity criterion is not of transcendental importance since several of the journals analyzed that are indexed do not meet it, the Redalyc database presents it as one of the highly valued criteria to be taken into account11.
Of the 55 journals analyzed, 40 (73%) meet the requirement of having at least 16 articles per year (Figure 3). At the same time, only 19 of the 55 journals (35%) showed homogeneity in the number of articles between the volumes of the last complete year of publication (Figure 3). The lack of homogeneity in terms of publications has a simple solution: to publish homogeneously. However, part of the sample of the journals analyzed happens that to reach the 16 articles per year, they have to balance the publications between issues, i.e., if in the first issue they presented 6 articles, to meet the annual minimum, they must present 10. Some of the editors interviewed confirmed that when initiating the indexing processes of the journal, they did so, taking into account these indicators and planning the issues around this criterion.
They are analyzing the comparison between private and public institutions to fulfill these criteria. The difference between articles per year is minimal between the two types of institutions. Regarding homogeneity of publications, public institutions have worse performance, with only 22% of their journals meeting this criterion compared to 46% of the private ones. This shows that the editorial line in private journals emphasizes homogeneity of publication per year compared to public journals.
Information Availability and Visibility
This segment is about the availability and visibility of the information on the platform. The fulfillment of these criteria allows users to have deep knowledge about the journal they are reading or submit articles. Digital preservation allows maintaining access to digital materials beyond technological changes12. This allows the prevalence of scientific articles for posterity. At the same time, the existence of digital preservation policies explicitly is an indication that allows journal users to detect whether the journal is predatory or not13. Hosting on electronic platforms independent from the university institution makes users interested in the journal arrive through a direct URL instead of entering from the university site, allowing faster and more efficient access. At the same time, the criteria: the presence of statistical data and distinction of current indexing records allow knowing both the statistics of the journal in terms of its performance and the quality of the journal since it allows knowing where it is indexed and the rank it occupies.
Regarding the criteria of digital preservation policy for documents and the use of independent digital platforms, compliance is very positive (Figure 4). This is seen with 43 out of 55 journals (78%) explicitly providing their digital document preservation policies and with 47 out of 55 journals (85%) complying with the use of independent digital platforms. Meanwhile, only 26 out of 55 journals (47%) show the use of some statistics on the web page and 32 out of 55 journals (58%) comply with the distinction of current records, i.e., to present explicitly, in detail and with links to the directories, databases and indexes in which they are found. Although compliance with the first two criteria is positive, some journals need to improve; however, compliance with statistics and the distinction of current records is low.
We can see varied differences when comparing public and private institutions with these criteria. Private institutions have better compliance in terms of the use of independent platforms, with 93% against 78% of the public ones and regarding the indicator of statistical data usage with 58% compliance against a low 37% by the public ones. In contrast, public institutions had a better percentage of compliance in presenting digital preservation policies for documents with 81% versus 75% for private institutions and with 63% of distinction of current records versus 54% for private institutions. The differences between the types of institutions are not significant.
Journal and author information
The last group of indicators, where the journal's criteria and author information are found, showed a percentage of non-compliance of more than 10% are those of the provision of information about the journal and the authors, visible in Figure 5. The DOI system identifies content objects in the digital environment with their names assigned to any entity for use in digital networks. It provides a simple method for accessing and reusing scholarly materials, facilitates data citation and thus increases the availability and recognition of research materials14. The use of ORCID, on the other hand, increases the discoverability of the authors' research results. It provides all the same information from works, awards and affiliations. This generates reliable profiles for researchers who publish in the journal and gives more credibility to the articles published before the readers15. In comparison, the use of bibliographic mastheads allows better identification of the particles present in the journal by having all the information available in this space.
As shown in Figure 5, compliance with the indicators: digital resource identifier and Bibliographic masthead are high. A total of 43 out of 55 journals (78%) comply with the use of DOI, while 45 out of 55 journals (81%) comply with the use of bibliographic mastheads. However, only 34 of 55 journals (62%) comply with author identifiers. While bibliographic mastheads are easy to resolve since it only involves adding them to the articles, the other two criteria are more complex. First, the DOI, the solution is simple, but it implies a monetary investment by the institutions since it is a paid service. At the same time, the use of identifiers by authors implies that they create the profiles and keep them updated.
Regarding compliance with author indicators, both types of institutions (public and private) are evenly matched, with private universities having 61% compliance and public universities 63% compliance. However, in the other indicators, it could be detected that private universities have a higher percentage of compliance, with 93% complying with the use of bibliographic headings and 82% complying with the use of DOI in comparison to 70% of public institutions complying with bibliographic headings and 74% with the use of DOI. These differences indicate that private universities have a slight advantage in managing journal and author information.
CONCLUSIONS
The main problems detected in the journals are the diversity of editors and authors. There are journals with editorial committees composed of editors from the same institution as the journal, which prevents them from being indexed in the Redalyc and Scielo databases. In terms of geographic diversity, the number of journals with editorial boards composed of editors from the same country is higher. This represents the main obstacle to progress indexing Scopus and Web of Science. As for the authors who publish in the journals, the panorama is the same as the editorial committees since they also present a high percentage of non-compliance.
The editorial directors of Ecuadorian journals interviewed for this research affirm that how they have managed to improve the diversity of editors and authors is based on the management of contacts or networking. The editorial committees were formed based on working relationships managed by the editors, who, through networks generated during their professional life, have links with researchers and academics with the necessary reputation and experience to be members of the editorial committee. At the same time, as the editorial board is being formed, the new editors can search for and suggest possible candidates be part of the journal in the future.
Regarding authors in journals, the editors interviewed present a scenario very similar to the situation mentioned for publishers. A large number of the articles published are through the networking of the journal editors, since this way they can attract articles of more significant impact and scientific relevance. As journals gain reputation based on the indexing achieved and their good practices, they also increase the number of articles they receive in calls for papers. However, to obtain more citations and a more significant impact, editorial boards make an intense evaluation of which articles to publish to achieve that goal. This does not mean they do not publish the articles submitted through calls for papers. These articles are reviewed in-depth. The editorial teams maintain constant communication with the authors to correct errors and improve the publication.
Compliance with the indicators regarding the type of institution (public or private) showed no marked difference between the two types of institutions.
RECOMMENDATIONS
It is recommended that Ecuadorian journals build and develop their journals according to the indexing criteria in the central databases since these are designed with the objective that the journals have a more significant impact.
For future research, it is suggested to analyze the bibliometric indicators of Ecuadorian journals to know the country's situation in terms of these indicators since they are also an essential factor in determining how to improve the positioning and scope of scientific journals.
Acknowledgments.
Special thanks to the ECOMS school of PUCESI for their constant support throughout the research process.
REFERENCES
1 Mendoza, S., & Paravic, T. (2006). Origen, clasificación y desafíos de las Revistas Científicas. Investigación y Postgrado, 49-75.
2 Rodríguez Muñoz, R., Socorro Castro, A., & Espinoza Cordero, C. (2019). Análisis de Scimago Journal & Country Rank,utilidad para el desarrollo bibliométrico en la Universidad Metropolitana del Ecuador. Publicando, 58-68.
3 Balladares-Burgos, J., García-Naranjo, Á., & Granda-Villamar, C. (2020). Perspectivas de la producción científica en las universidades del Ecuador. Revista Cátedra, 3(2), 126-149.
4 Simbaña Gallardo, V. P., Espinosa Carrera, K. S., & Vinueza Castillo, D. E. (2020). Perspectivas a la producción científica del Ecuador. Tsafiqui - Revista Científica En Ciencias Sociales, 11(14), 117-129. doi:https://doi.org/10.29019/tsafiqui.v14i1.656
5 Garcia-Bereguiain, M. M. (2019). Revistas Indexadas en Scopus en Ecuador: La Asignatura Pendiente. Revista Ecuatoriana de Neurología, 13-14.
6 SCImago. (2020). SJR — SCImago Journal & Country Rank. Obtenido de http://www.scimagojr.com
7 Goyanes, M., & Demeter, M. (2020). How the Geographic Diversity of Editorial Boards Affects What Is Published in JCR-Ranked Communication Journals. Journalism & Mass Communication Quarterly, 97(4), 1123-1148. doi:10.1177/1077699020904169
8 Ribera, M., Pozzobon, R., & Sayago, S. (2019). Publishing accessible proceedings: the DSAI 2016 case study. Universal Access in the Information Society, 19(3), 557-569. doi:10.1007/s10209-019-00660-3
9 Márquez, M., & Porras, A. (2020). Science Communication in Multiple Languages Is Critical to Its Effectiveness. Frontiers in Communication, 5. doi:10.3389/fcomm.2020.00031
10 Ansorge, L. (s.f.). Let's publish full-text scientific articles in HTML, not just PDF. European Science Editing. doi:10.3897/ese.2021.e75834
11 Redalyc. (2020). Postula tu revista. Recuperado el 5 de 12 de 2021, de https://www.redalyc.org/postulacion.oa?q=criterios
12 Saini, O. (2018). Understanding the Role of Institutional Repository in Digital Preservation in Academic Libraries: Review of Literature. Library Philosophy and Practice (e-journal), 1904. Obtenido de https://digitalcommons.unl.edu/libphilprac/1904/?utm_source=digitalcommons.unl.edu%2Flibphilprac%2F1904&utm_medium=PDF&utm_campaign=PDFCoverPages
13 Laine, C., & Winker, M. (2017). Identifying predatory or pseudo-journals. Biochemia Medica, 27(2), 285-291. doi:10.11613/bm.2017.031
14 Wang, W., Deng, L., You, B., Zhang, P., & Chen, Y. (2017). Digital object identifier and its use in core Chinese academic journals: A Chinese perspective. Learned Publishing, 31(2), 149-154. doi:10.1002/leap.1137
15 Meadows, A. (25 de 08 de 2017). Ten reasons to get — and use — an ORCID iD! Obtenido de Elsevier Connect: https://www.elsevier.com/connect/authors-update/ten-reasons-to-get-and-use-an-orcid-id!
Received: 24 December 2021 / Accepted: 1 February 2022 / Published: date. 15 february 2022
Citation: Santiago Padilla V, Ramos Gil Y T. Indicators to achieve a better positioning of Ecuadorian scientific journals. Revis Bionatura 2022;7(1). 2. http://dx.doi.org/10.21931/RB/2022.07.01.2
