2019.04.02.3
Files > Volume 4 > Vol 4 No 2 2019
INVESTIGATION / RESEARCH
Polypharmacological study of Ceritinib using a structure based in silico approach
Tammanna R. Sahrawat1 and Prabhjeet Kaur2
Available from: http://dx.doi.org/10.21931/RB/2019.04.02.3
ABSTRACT
Drug repurposing has gained mass recognition over the past few years as it has paved new therapeutic applications for already approved FDA drugs. It focuses on finding new molecular targets of drugs for medical uses different than the one originally proposed. Ceritinib, an Anaplastic Lymphoma Kinase (ALK) inhibitor is given orally in the treatment of non-small cell lung cancer (NSCLC). This treatment has been reported to be associated with a number of side effects such as hyperglycemia, convulsion, pneumonitis etc. The side effects are usually due to the unintended interaction of the drug with other protein targets. In silico polypharmacological studies of Ceritinib suggests that it binds to multiple targets other than the intended one which may largely be due to different proteins possessing similar binding sites. ProBis server was used to retrieve probable off-targets of Ceritinib based on presence of structurally similar protein binding sites as that of ALK. Ceritinib was found to bind effectively to three proteins namely Lymphocyte Cell-Specific Protein-Tyrosine Kinase, Tropomyosin receptor kinase B and Aurora kinase B having favorable binding energies and inhibition constants, with no reported side-effects as compared to their marketed drugs. Therefore, it is concluded from the present study that Ceritinib may act as an effective therapeutic target against its polypharmacological targets.
Keywords: Polypharmacology, repurposing, molecular docking, protein binding similarity search.
INTRODUCTION
Conventional drug discovery processes require huge time and a hefty amount of money to bring a single drug into the market. Hence, in silico methods play a vital role in drug discovery and development by offering an enhanced efficiency to the pharmaceutical industry1. Over the last four decades, the drug manufacturing companies have shifted their focus from the reductionist approach towards ‘one drug, multiple targets’ also known as polypharmacology. Polypharmacology has been defined as the ability of small molecules to interact simultaneously and specifically with multiple targets2. Polypharmacology can be broadly characterized into two types: adverse polypharmacology (which involves off–target binding) and therapeutic polypharmacology (which involves repurposing of drugs i.e. the process of searching new therapeutic applications for existing drugs also known as drug repositioning)3.
Polypharmacology thus offers to be a promising approach for identifying novel therapeutic uses for already known drugs against complex diseases. It has been previously reported that effect of a drug on an unintended target might suggest new uses for an existing drug (i.e. multi-target drugs)4. Moreover, drugs which failed to provide the intended benefit can also be considered for repurposing for the treatment of other diseases. A classic example is the anti-cancer drug Zidovudine which failed in intended treatment of cancer however it was found to be effective against HIV and in 1987 was approved by FDA for HIV treatment5.
Polypharmacological drugs are expected to have relatively lower chances for developing drug resistance as they target multiple drug targets and are considered to show better efficacy than drugs for a single target6,7. Several drugs have already been successfully repurposed for new diseases, outside the original medical scope. For example, the blockbuster drug Sildenafil (Viagra), a phosphodiesterase (PDE) inhibitor was first developed for treating hypertension and ischemic heart disease. During the phase I clinical trials, Sildenafil was found to induce penile erections as a side effect and after phase II clinical trial failure, sildenafil was repositioned for the treatment of erectile dysfunction and approved by the FDA8.
Ceritinib (formerly known as LDK378 and marketed as Zykadia™), an inhibitor of Anaplastic lymphoma kinase is used to treat non-small cell lung cancer (NSCLC). It acts to inhibit the autophosphorylation of Anaplastic Lymphoma Kinase (ALK) and its downstream signaling protein STAT3 thereby preventing the proliferation of ALK-dependent cancer cells9.
In silico computational methods are of high biological and pharmacological value as they predict new targets for existing small-molecule compounds2. In our previous in silico polypharmacological study, it was found that hyperglycemia caused in patients after Ceritinib treatment could be due to the off-target binding of Ceritinib to dual specificity protein kinase CDC Like Kinase 2 (CLK2) (PDB ID: 3NR9) along with its actual target Anaplastic Lymphoma Kinase10. Therefore, in the present study, we further investigated the polypharmacological-targets of ceritinib to identify its probable repurposing potential.
MATERIAL AND METHODS
Data mining of target protein of Ceritinib
The Drugs and Drug Targets mapping tool of the Protein Data Bank (PDB) was utilized to obtain the crystal structure of the target protein Anaplastic lymphoma tyrosine kinase (ALK) bound with Ceritinib (PDB ID: 4MKC)11.
Identification of proteins with similar binding sites
The ProBis server was used to detect structurally similar protein binding sites in ALK tyrosine kinase (PDB ID: 4MKC). ProBis employs a fast maximum clique algorithm to detect similar protein binding sites independent of the sequence or the fold present in proteins12. Therefore, using ProBis proteins having similar geometrical and physicochemical properties to Ceritinib target ALK were retrieved.
Docking studies of Ceritinib with probable polypharmacological/off-targets
To investigate the binding affinity of Ceritinib with proteins other than its actual target ALK tyrosine kinase, Autodock tool was used to perform drug-protein docking13. The binding energies and inhibition constants of polypharmacological-targets with Ceritinib and their bound ligands were tabulated and compared.
Literature mining
The probable polypharmacological-targets of Ceritinib were studied for their role in disease pathways and therapeutics in order to predict its repurposing prospects.
RESULTS AND DISCUSSION
Repurposing studies involve investigating multiple targets of drugs to enhance their biological activities as well as reduce their toxicities. In order to find polypharmacological targets of Ceritinib, the drug was docked with 33 probable off-targets obtained from ProBis server. Literature studies also revealed that these polypharmacological targets may be critical therapeutic targets in various other complex diseases such as breast cancer, Acute Lymphoblastic Leukemia (ALL) and CNS disorders. Sahrawat and Kaur (2018) while investigating the polypharmacology of Certinib have reported that proteins having similar binding sites to the actual target of Certinib i.e. ALK , retrieved from ProBis server, namely Ribosomal protein S6 kinase alpha-1 (RSK1) (PDB ID: 2Z7R), Lymphocyte Cell-Specific Protein-Tyrosine Kinase (LCK) (PDB ID: 1QPC), Tropomyosin receptor kinase B (TRKB) (PDB ID: 4AT5) and Aurora kinase B (AURKB) (PDB ID: 4AF3) had a significant Z-score > 2.0. Z-score is the measure of similarity between two protein binding sites12 due to which different proteins may interact with same drug molecules. Further, the docking analysis of these proteins with the drug Ceritinib, revealed binding energies and inhibition constants comparable to the actual target i.e. ALK, with no reported side-effects on binding to Ceritinib10. Therefore, the proteins which were identified as off-targets of Ceritinib, were further investigated for repurposing of Ceritinib, so as to identify other probable usage of the drug.
The results obtained from docking server, Autodock is in terms of binding energy and inhibition constant (Ki) at temperature 298.15 K. Binding energy is the affinity of the ligand with which it binds to the target protein. More negative binding energy, higher is the affinity of the ligand to bind the receptor protein. Inhibition constant of a drug is the concentration required to produce half maximum inhibition on binding the receptor, therefore, lower the inhibition constant value lesser is the amount of drug concentration needed to produce an effect13.
The docking studies indicated that the binding energy of Ceritinib with its actual target, ALK (PDB ID: 4MKC) was -5.62 kcal/mol with an inhibition constant of 75.9. In addition, the docking studies of Ceritinib with proteins Ribosomal protein S6 kinase alpha-1 (RSK1) (PDB ID: 2Z7R), Lymphocyte Cell-Specific Protein-Tyrosine Kinase (LCK) (PDB ID: 1QPC), Tropomyosin receptor kinase B (TRKB) (PDB ID: 4AT5) and Aurora kinase B (AURKB) (PDB ID: 4AF3) revealed that they had higher binding affinities and lower inhibition constant for Ceritinib than its actual target ALK (PDB ID: 4KMC)10 as shown in Table 1.
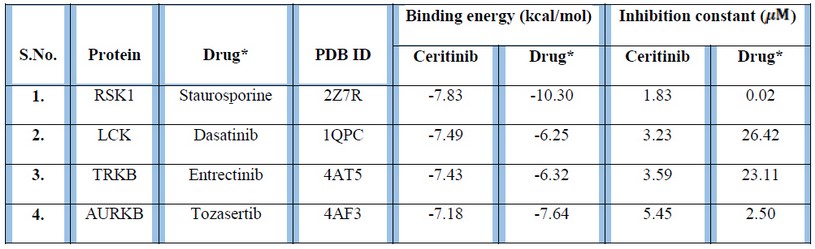
Table 1: Binding energy and inhibition constant values of polypharmacological-targets with Ceritinib and their reported therapeutic agents.
Ribosomal protein S6 Kinase alpha-1 (PDB ID: 2Z7R) retrieved from ProBis server had binding energy of -7.83 kcal/mol with Ceritinib while its marketed therapeutic agent Staurosporine (Drugbank ID: DB02010) had a higher binding energy of -10.3 kcal/mol. Further, RSK1 had a favorable inhibition constant for Staurosoporine, which has been used for treating triple negative breast cancer14,15 (Table 1). Our results are also supported by the report of Kuenzi et al. (2017) in an integrated functional proteomics study for repurposing of the anaplastic lymphoma kinase (ALK) inhibitor Ceritinib, wherein it was reported that IGF1R, RSK1, RSK2 and FAK1 are potential off-targets of Ceritinib16.
LCK is a potential target in the treatment of T-cell Acute Lymphoblastic Leukemia (ALL) and its marketed drug is Dasatinib (D number: D03658)17, 18. Moreover, for the treatment of Central nervous system (CNS) disorders and various tumors including NSCLC, TRKB has been used as a potential therapeutic target and Entrectinib (D number: D10926) is prescribed for its treatment19,20. The probable polypharmacological targets of Ceritinib, namely LCK protein (PDB ID: 1QPC) and TRKB (PDB ID: 4AT5) had higher binding affinity and lower inhibition constant for Ceritinib as compared to their marketed therapeutic agents Dasatinib and Entrectinib, respectively (Table 1).
On docking of Aurora kinase B (PDB ID: 4AF3) with its marketed therapeutic agent Tozasertib (D number: D08279)21, it was found that there was not much difference in their binding energies, although the inhibition constant was higher for Ceritinib. AURKB has been reported to be involved in various cancers such as breast cancer, hepatocellular carcinoma, lung adenocarcinoma, colorectal cancer models and acute lymphoblastic leukemia (ALL) which are currently being treated using Tozasertib22,23.
On investigation of binding site residues of polypharmacological-targets of Ceritinib it was seen that all the proteins RSK1, LCK, TRKB and AURKB had similar binding site interacting residues as that of Ceritinib with its actual target ALK (PDB ID: 4KMC) tabulated in Table 2, Figure 1.
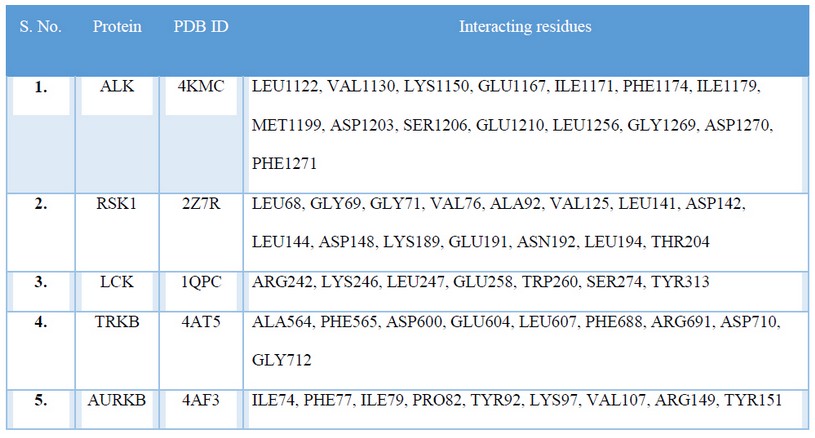
Table 2: Binding site residues of actual target and polypharmacological-targets of Ceritinib.
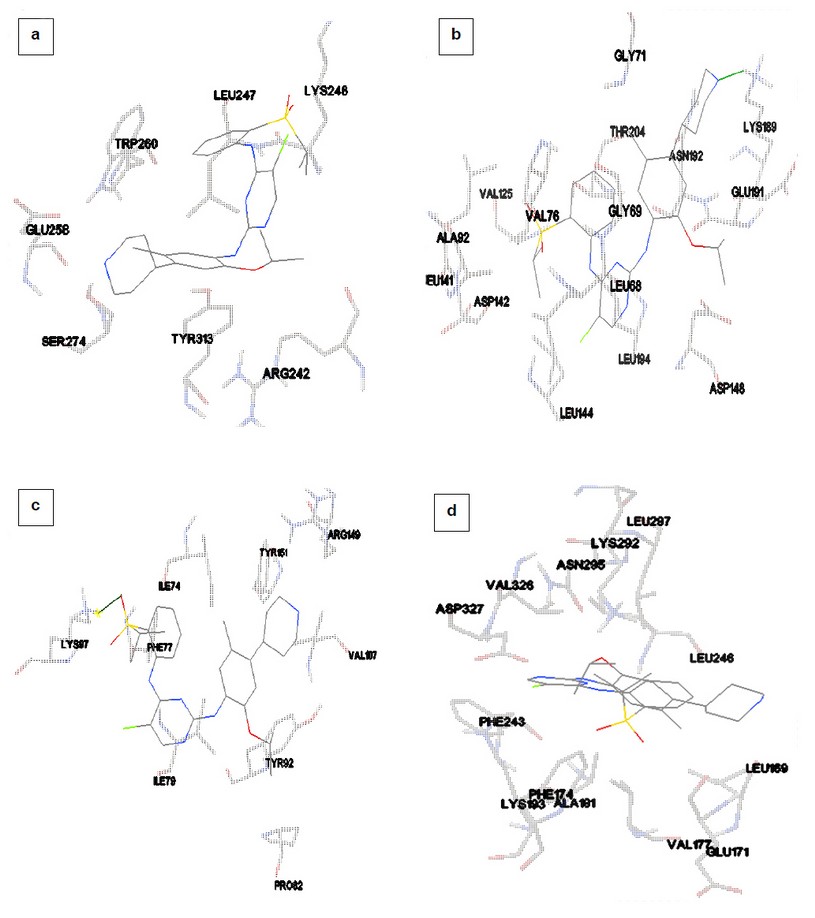
Figure 1: Interacting amino acid residues in the binding pockets of a) Ribosomal protein S6 Kinase alpha 1 (PDB ID: 2Z7R) b) LCK (PDB ID: 1QPC) c) TRKB/BDNF/NT-3 growth factors receptor (PDB ID: 4AT5) d) Aurora Kinase B (PDB ID: 4AF3) docked with Ceritinib visualized using AutoDock analysis tool.
Analysis of the docking scores of all the four poypharmacological targets namely Ribosomal protein S6 kinase alpha-1, LCK, Aurora kinase B and (Table 2) indicated that Ceritinib binds effectively to three proteins namely LCK, AURKB and TRKB as compared to their available marketed drugs.
CONCLUSION
Therefore, it may be concluded that three polypharmacological targets of Ceritinib i.e LCK, Aurora kinase B and Tropomyosin receptor kinase B, identified in the present study, bind to the Ceritinib more effectively, as seen from their high binding energies, as compared to the drug’s actual target ALK. This study paves new opportunities for Ceritinib in the treatment of various complex diseases such as cancer, central nervous disorders etc. associated with these proteins. In addition, experimental studies need to be carried out to further validate the results of this work.
Acknowledgement
The authors would like to thank their parent institute Panjab University, Chandigarh, India for providing the infrastructure for carrying out the research.
REFERENCES
1. Kumar, N., Hendriks, B. S., Janes, K. A., de Graaf, D., & Lauffenburger, D. A. (2006). Applying computational modeling to drug discovery and development. Drug discovery today, 11(17-18), 806-811.
2. Rastelli, G., & Pinzi, L. (2015). Computational polypharmacology comes of age. Frontiers in pharmacology, 6, 157.
3. Boran, A. D., & Iyengar, R. (2010). Systems approaches to polypharmacology and drug discovery. Current opinion in drug discovery & development, 13(3), 297.
4. Ashburn, T. T., & Thor, K. B. (2004). Drug repositioning: identifying and developing new uses for existing drugs. Nature reviews Drug discovery, 3(8), 673.
5. Broder, S. (2010). The development of antiretroviral therapy and its impact on the HIV-1/AIDS pandemic. Antiviral research, 85(1), 1-18.
6. Anighoro, A., Bajorath, J., & Rastelli, G. (2014). Polypharmacology: challenges and opportunities in drug discovery: miniperspective. Journal of medicinal chemistry, 57(19), 7874-7887.
7. Peters, J. U. (2013). Polypharmacology–foe or friend?. Journal of medicinal chemistry, 56(22), 8955-8971.
8. DeBusk, R. F., Pepine, C. J., Glasser, D. B., Shpilsky, A., DeRiesthal, H., & Sweeney, M. (2004). Efficacy and safety of sildenafil citrate in men with erectile dysfunction and stable coronary artery disease. American Journal of Cardiology, 93(2), 147-153.
9. Chen, Z., Fillmore, C. M., Hammerman, P. S., Kim, C. F., & Wong, K. K. (2014). Non-small-cell lung cancers: a heterogeneous set of diseases. Nature Reviews Cancer, 14(8), 535.
10. Sahrawat, T. R., & Kaur, P. (2018). Investigation of Off-targets of NSCLC drug Ceritinib: A Structure Based Systems Biology Approach. International Journal of Advanced Scientific Research and Management (IJASRM), 3(12), 31-36.
11. Sussman, J. L., Lin, D., Jiang, J., Manning, N. O., Prilusky, J., Ritter, O., & Abola, E. E. (1998). Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallographica Section D: Biological Crystallography, 54(6), 1078-1084.
12. Konc, J., & Janežič, D. (2012). ProBiS-2012: web server and web services for detection of structurally similar binding sites in proteins. Nucleic acids research, 40(W1), W214-W221.
13. Morris, G. M., Huey, R., Lindstrom, W., Sanner, M. F., Belew, R. K., Goodsell, D. S., & Olson, A. J. (2009). AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. Journal of computational chemistry, 30(16), 2785-2791.
14. Xue, L. Y., Chiu, S. M., & Oleinick, N. L. (2003). Staurosporine-induced death of MCF-7 human breast cancer cells: a distinction between caspase-3-dependent steps of apoptosis and the critical lethal lesions. Experimental cell research, 283(2), 135-145.
15. Stratford, A. L., Reipas, K., Hu, K., Fotovati, A., Brough, R., Frankum, J.,& Lasham, A. (2012). Targeting p90 ribosomal S6 kinase eliminates tumor‐initiating cells by inactivating Y‐box binding protein‐1 in triple‐negative breast cancers. Stem cells, 30(7), 1338-1348.
16. Kuenzi, B. M., Rix, L. L. R., Stewart, P. A., Fang, B., Kinose, F., Bryant, A. T., & Rix, U. (2017). Polypharmacology-based ceritinib repurposing using integrated functional proteomics. Nature chemical biology, 13(12), 1222.
17. De Keersmaecker, K., Porcu, M., Cox, L., Girardi, T., Vandepoel, R., de Beeck, J. O., & Hantschel, O. (2013). NUP214-ABL1 mediated cell proliferation in T-cell acute lymphoblastic leukemia is dependent on the LCK kinase and various interacting proteins. haematologica, haematol-2013.
18. Ottmann, O., Dombret, H., Martinelli, G., Simonsson, B., Guilhot, F., Larson, R. A., & Gollerkeri, A. (2007). Dasatinib induces rapid hematologic and cytogenetic responses in adult patients with Philadelphia chromosome–positive acute lymphoblastic leukemia with resistance or intolerance to imatinib: interim results of a phase 2 study. Blood, 110(7), 2309-2315.
19. Doebele, R. C., Davis, L. E., Vaishnavi, A., Le, A. T., Estrada-Bernal, A., Keysar, S., & Stephens, P. J. (2015). An oncogenic NTRK fusion in a patient with soft-tissue sarcoma with response to the tropomyosin-related kinase inhibitor LOXO-101. Cancer discovery, 5(10), 1049-1057.
20. Rolfo, C., Ruiz, R., Giovannetti, E., Gil-Bazo, I., Russo, A., Passiglia, F., & Raez, L. (2015). Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert opinion on investigational drugs, 24(11), 1493-1500.
21. Harrington, E. A., Bebbington, D., Moore, J., Rasmussen, R. K., Ajose-Adeogun, A. O., Nakayama, T., & Su, M. (2004). VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nature medicine, 10(3), 262.
22. Tang, A., Gao, K., Chu, L., Zhang, R., Yang, J., & Zheng, J. (2017). Aurora kinases: novel therapy targets in cancers. Oncotarget, 8(14), 23937.
23. Hartsink-Segers, S. A., Zwaan, C. M., Exalto, C., Luijendijk, M. W. J., Calvert, V. S., Petricoin, E. F.,& Hansen, B. R. (2013). Aurora kinases in childhood acute leukemia: the promise of aurora B as therapeutic target. Leukemia, 27(3), 560.
Received: 24 April 2019
Accepted. 10 May 2019
Tammanna R. Sahrawat1 and Prabhjeet Kaur2
1Assistant Professor, 2Post-graduate student
1, 2Centre for Systems Biology & Bioinformatics, UIEAST
Panjab University, Chandigarh, India
Corresponding Author: Dr. Tammanna R. Sahrawat* Ph.D.
Email: [email protected]
