2023.08.02.24
Files > Volume 8 > Vol 8 No 2 2023
16SrRNA sequencing analysis for identification of Klebsiella pneumoniae isolated from the extreme kitchen environment
1Department of Biology, College of Science, Mosul University, Mosul, Iraq; [email protected]
2Department of Biology, College of Science, Mosul University, Mosul, Iraq;. E-mail: [email protected]
2,* Department of Biology, College of Science, Mosul University, Mosul, Iraq;. E-mail: [email protected]
* Correspondence: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.02.24
Swabs from dishwasher samples were collected and cultured on different media, and then a gram stain was conducted on pure colonies to find whether they were Gram-positive or negative. 32-gram negative Isolates were obtained from the dishwasher; then, we chose isolates under study depending on morphological features on previous media for further investigation. 32 Gram-negative isolates were obtained from a dishwasher, and three isolates of Klebsiella pneumoniae were diagnosed by some phenotypic characteristics and approved by using 16 SrRNA partial sequencing analyses. The 3 isolates deposited in the NCBI database under accession number OK 254156.1 for K.pneumoniae strain NPK 323, OK 247423.1 for K. pneumonia as strain CUMB SAM-61, and OK245427.1 for K. pneumonia strain PD17. The phylogenetic tree for 3 isolates was done by using MEGA II software. Many experiments have been conducted on two isolates (OK 247423.1 and OK245427.1 ) because the OK 254156.1 strain was lost during laboratory work and repeated cultures. A hemolysis test on blood agar and a lipase test on egg–yolk agar were done; both isolates showed negative results for hemolysis blood and producing lipase enzyme, while both isolates showed their ability to produce lecithinase enzyme. The two isolates gave an excellent result in the tube method for the biofilm formation test. Also, a good candidate production test was obtained for these two isolates using L.B. acetate agar medium. Conclusion: Bacterial species differ according to the environments in which they live, as the species that are isolated from clinical sources and possess many virulence factors that make them more dangerous and pathogenic to humans differ about the same species if isolated from a variety of external environments, which makes them virulent or have new characteristics that make them adapted to live in the environments from which they are isolated.
Keywords: Klebsiella pneumonia, virulence factor, extreme conditions, phylogenetic tree.
INTRODUCTION
Scientists are always trying in their microbial biological research and studies to search for species that can withstand harsh environmental conditions, whether in water, air or soil. Europe finally discovered several hardy bacteria and fungi lurking on the rubber seals of dishwasher doors, some of which are opportunistic pathogens. These microbes must withstand high temperatures, high NacL Concentration, acidity, cleaning products, and the shear force of water during washing cycles to establish a home there1.
Studying bacterial diversity that has survived in complex environments has contributed to understanding how life has developed. Despite their limited worldwide distribution, severe environments are home to many microorganisms important to science and technology2, such as bacteria producing antibiotics3 and those involved in pollution biodegradation4. Many taxa are represented in these habitats, giving a wealth of knowledge on microbial physiology and biodiversity.
Gram-negative bacilli are characterized by several characteristics, including that they are widespread in environmental systems, where they are found in air, water, and soil, in addition to their presence in sewage systems and wet places. What also distinguishes them is that they need few nutrients for growth, unlike other species, and they are characterized by resistance to chemical antibiotics5,6,7.
Klebsiella bacteria are characterized by causing many respiratory system diseases, infecting the blood circulation of the heart, and causing severe infections in hospitals. This pathogen is also one of the most commonly isolated bugs in community-acquired illnesses.. The gastrointestinal system may be the primary pathogenic reservoir for transmitting these dangerous diseases, with the majority of cases coming from the hands of hospital staff or healthcare workers. Klebsiella pneumoniae is an opportunistic pathogen that targets individuals whose immune has been weakened by an underlying disease such as diabetes mellitus, persistent lung obstruction, or major surgery; also often detected in inhalation; in addition, their clinical relevance in individuals whose immune is weakened by underlying disease is challenging to determine8,9,10,11,12.
Isolation
Swabs from dishwasher samples were collected by passing sterile swabs on the specific area in 6 dishwashers. The swabs were cultured on different media such as Nutrient agar, MacConkey agar, Blood agar and methylene blue agar and incubated at 37C for 24 hours, when isolated and cultured continually until they were obtained pure colonies. Then gram stain was conducted on pure colonies to find whereas Gram-positive or negative. 32-gram negative Isolates were obtained from the dishwasher; then, we chose isolates under study depending on morphological features on previous media for further investigation.
Identification by 16SrRNA sequencing:
Three isolates of Klebsiella were selected for identification by 16SrRNA.In the first step of identification, the total DNA of isolates is extracted depending on the instruction of a specific kit (G-spin DNA kit, intron biotechnology ). The primers of the PCR reaction included forward primer (5'- AGAGTTTGATCCTGGCTCAG- 3') and reverse primer(5'- GGTTACCTTGTTACGACTT- 3’), the final volume of the reaction was done as 25 ML that is composed of 5ML of pre mix,1ML from each forward primer and reverse primer, and then added 1.5 ML of total DNA.finally the total volume of reaction completed by 16.5 ML of D.W into 25ML. The condition of the reaction is in Table 1.
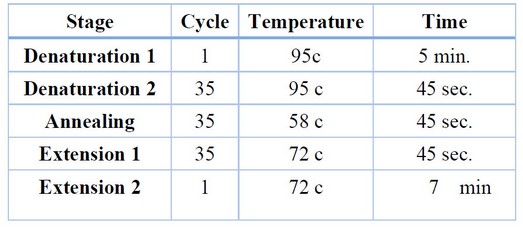
Table 1. The reaction of PCR.
Phylogenetic tree analysis
The genealogical tree method was used to trace bacterial species and genera through their genetic fingerprints and the composition of the DNA to its nucleotides. There were a total of 1485 positions in the final dataset. Evolutionary analyses were conducted in MEGA11
Detection of Virulence Factors of K. pneumoniae Isolates
Hemolysin test
A hemolysin test was carried out to determine the hemolytic activity of Klebsiella pneumoniae isolates; two Test isolates were inoculated in a blood agar plate that contained 5%(vol\vol) of human blood after activated in nutrient broth for incubation at 37°C, for 24 hours. The presence or absence of hemolysis in the surrounding colonies was recorded13.
Production of lecithinase and lipase enzyme:
By using egg–yolk agar medium, the ability of the isolates was tested for the production of lecithinase and lipase enzyme; the medium contained 85 ml of nutrient agar and 15 ml of egg-yolk, sterilized the nutrient agar and then cool to 55c added egg –yolk and mixed then poured in plates.
The isolates were inoculated in the plate and incubated at 37C for 24h. The plates were filled with a saturated copper sulfate aqueous solution and let to stand for 20 minutes before the surplus solution was drained. The results were read after a few minutes of drying. The green-blue color of opalesces showed lipolysis synthesis, and the area of clearing surrounding the colony was declared positive for lecithinase production14,15,16.
Biofilm formation:
The two bacterial isolates were screened for the ability of biofilm(method) formation by using the test tube method. In the test tube method, the isolates were grown in the test tube containing tryptic soy broth and 1% glucose and incubated for 24h at 37C; then, the media was discarded. The lines were gently rinsed with phosphate buffer saline and left to dry, then 1ml of 1% w/v crystal violet solution was added to the tube to stain the cells attached to the walls, Crystal violet solution was discarded after 3 min. The tubes were washed with deionized water and left to dry inverted. The staining of the walls of tube 17 indicated biofilm synthesis.
Investigation of the ability of Klebsiella pneumonia to produce calcite:
Klebsiella pneumonia isolates were grown on Luria-Bertani acetate agar medium (Tryptophane 1g, Yeast extract 0.5g. Nacl 0.05g, Calcium acetate 1g, Agar 1.5g, D.W 100ml)the Ph adjusted at (8) and incubated at 37C for a week. After the incubation period was over, the formation of calcite crystals was investigated18
Chemical diagnosis of calcite crystals:
A chemical examination of the crystals was carried out by adding drops of HCL 10% solution to the calcifications to observe the formation of bubbles as a result of the formation of carbon dioxide gas as a product of the reaction, and the result was recorded19
The isolates under study showed coccobacilli gram-negative cells, and the morphological features on McConkey agar showed 3 high mucus strains with pink mucoid colonies, while on Eosin methylene blue agar are appear pink to purple colonies finally, colonies on blood agar are smooth raised mucoid colonies. figure (1)
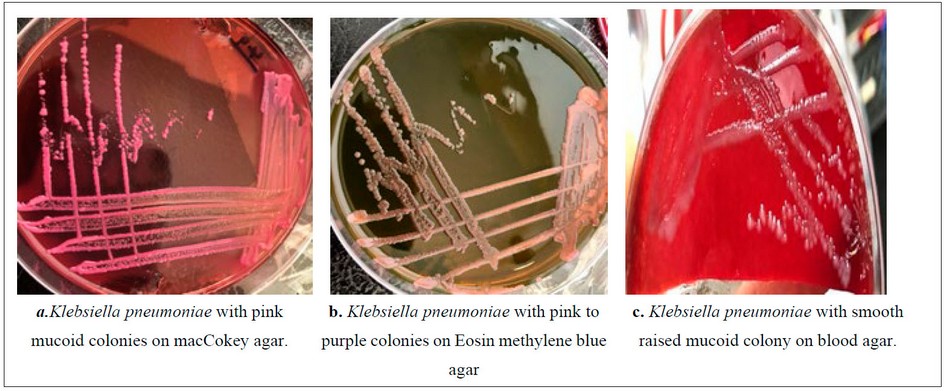
Figure. 1. Klebsiella pneumoniae on different media
The 3 mucus strains were selected for 16SrRNA sequencing analysis by amplifying 16S rRNA genes and analysis by electrophoresis on 1% agarose gel, as shown in Figure ( 2 ).
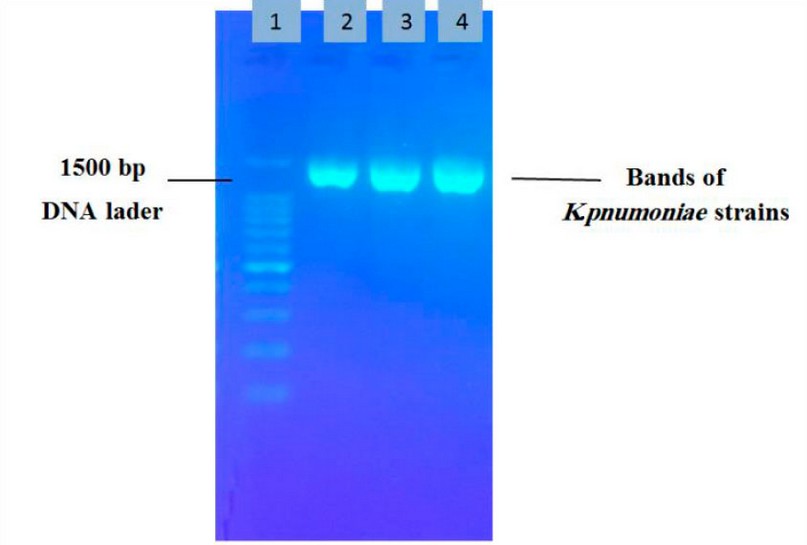
Figure 2. Electrophoresis of 16SrRNA products by agarose gel 1:DNA ladder , 2:OK247423, 3:OK254156 , 4:OK254156
The result of 16SrRNA sequencing compared with NSBI database, through NSBI blast with similarity 99%of standard K.pneumoniae isolates as (K. pneumoniae NPK 323, K. pneumoniae CUMB SAM-61 and K. pneumonia PD17).
The use of 16SrRNA analysis is regarded as an appropriate tool for the specific identification of isolates under study and gives a proven method for molecular analysis. The 3 isolates of K. pneumoniae were deposited in NSBI with accession numbers OK 254156.1 for K.pneumoniae strain NPK 323, OK 247423.1 for K. pneumoniae as strain CUMB SAM-61, and OK245427.1 for K. pneumonia strain PD17.
The phylogenetic analysis done by the Neighbor-Joining tree showed a relationship between OK 247423.1 CUMB SAM—61 and OK254156.1 NPK 323 with other K.pneumoniae strains recently deposited in NSBI database (The OK247423.1 and OK254156.1 strains) showed related to MH938261.1 strain. Still, the relationship between OK247423.1 and MH938261.1 was more closely. the strain OK245427 was closely related to the KU711913 strain (figure 3).
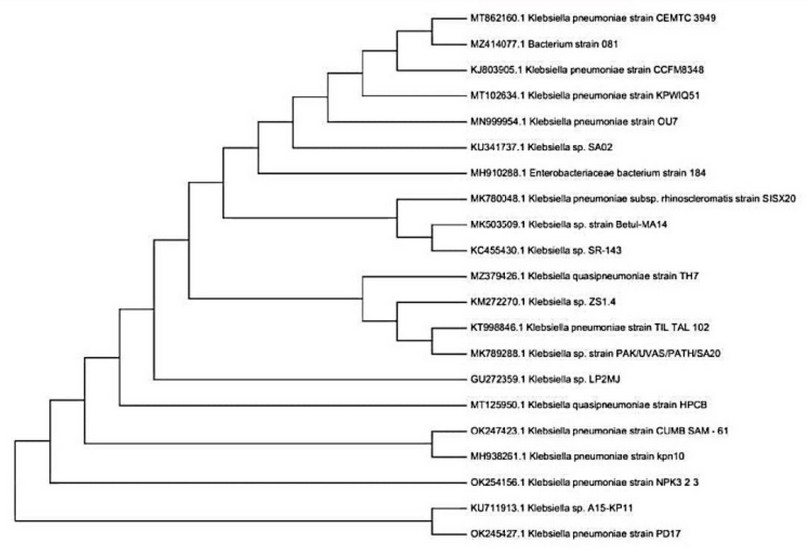
Figure 3. Phylogenetic tree.
Klebsiella pneumoniae isolates and their ability for hemolysis blood, production of lecithinase and lipase enzymes, biofilm and calcite formation:
The two isolates of Klebsiella pneumonia ((OK 247423.1 and OK245427.1 )) give a negative result for hemolytic activity on blood agar plates, and the same two isolates of Klebsiella pneumoniae showed their ability for lecithinase production by formation a clear zone around the bacterial colonies, as shown in (figure 4). In contrast, it gives a negative result with lipase enzymes.

Figure 4. Klebsiella pneumoniae on egg yolk agar with clear zone around colonies.
Our findings refer to the two isolates of Klebsiella pneumonia((OK 247423.1 and OK245427.1 )) which showed the ability for biofilm formation on the inner walls and down of tubes in the form of a violet color, as shown in (figure 5). While L. B. acetate agar showed calcite formation through the calcifications on the surface of the growing colonies, as shown in (figure 6).

Figure 5. formation of biofilm by Klebsiella pneumoniae
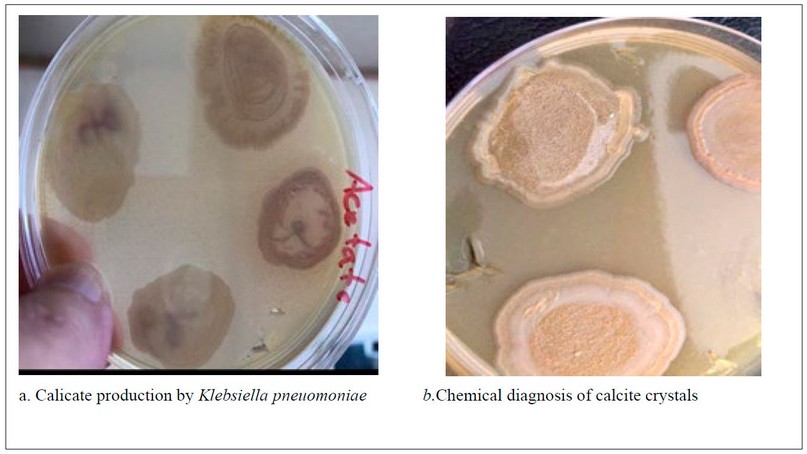
Figure 6. Klebsiella pneuomoniae on L.B. acetate agar
DISCUSSION
Life evolution produced new chances for microbial development, resulting in a shift in the microorganisms we encounter20.
Microorganisms enter dwellings with air, soil, water, live plants, various food products, other pets, and humans21,22. Most available overviews of indoor microbiomes focus on air-borne, dust-associated microbes23. Compared with few published research on microbes in humid household environments such as bathrooms, kitchens, and appliances that need water in their work.
The isolates of Klebsiella pneumoniae obtained from extreme environments differ in their possession of virulence factors. Our isolates could not cause hemolysis of blood, and this result agrees with Pereira and Vanetti24 may be due to the absence of activators for gene expression that code for this enzyme25. In recent research, none of the 54 strains isolated had blood hemolysis or possessed the hlyA and cnf-1 genes, which code for this activity. Although hemolytic activity has been documented, few studies show the genotypic and phenotypic existence of hemolysis in Klebsiella25,26. At the same time, our result differs from Kalaivani and his group27, who reported that the Klebsiella pneumonia isolates had a high percentage of hemolytic activity.
The two isolates of Klebsiella pneumoniae showed their ability for lecithinase production by forming a large zone of opalescence around the colonies. At the same time, give a negative result with the production of lipase enzyme. Studies have varied concerning the output of lecithinase enzyme. This research by Singh28, with 208 Klebsiella pneumoniae isolates (7.69%), produced lecithinase activity. And a research study by Kalaivani,55.7% (64) Klebsiella pneumoniae isolates observed Lecithinase production, which might predict more pathogenic ability among this isolates27 Our result differs from other study29 which referred that Klebsiella pneumoniae isolates, cannot produce lecithinase enzyme. At the same time, the development of our study agrees with the same study in the inability of Klebsiella pneumoniae isolates to produce lipase enzyme (figure 4).
Microbial biofilms are colonies of clustered microbial cells immersed in an extracellular polymeric material matrix that they generate by themselves (EPS). Biofilms can protect bacteria against ultraviolet (U.V.) radiation,, high pH, severe temperature salt stress, high pressure, inadequate nutrition, antibiotics, and other factors by functioning as "protective clothing."30
Our finding refer the two isolates of Klebsiella pneumoniae that showed good ability for biofilm formation, and this agreement with31which they found that 93.6% of K. pneumoniae isolates formed biofilms. These strains were divided into four categories as strains that generate a moderate biofilm, completely established biofilms, insignificant biofilms, and non-biofilm providers.
Also, our result was similar to 27, found among the several virulence strategies investigated, mainly of Klebsiella pneumoniae isolates (79%) demonstrated biofilm methods. In another recent study, 100 % biofilm production was documented for Klebsiella pneumoniae isolates32., while the study of Abbas in 2020 33 showed that only 4 out of 73 isolates of Klebsiella pneumoniae were able to form biofilms (figure 5).
Microbially induced calcite precipitation (MICP) is the generation of calcium carbonate from a saturated solution caused by microbial cells and metabolic activity 24. During MICP, organisms can emit one or more metabolic products (CO3 2), which react with ions (Ca2+) in the environment, resulting in mineral precipitation.
The MICP process is an eco-friendly and effective technology that may be used to solve a variety of environmental concerns, including radionuclide cleanup, heavy metal remediation, consolidation, CO2 sequestration, bio cement,, and other uses35
The results of two Klebsiella pneumonia isolates on L. B. acetate agar showed calcite formation by creating calcifications on the surface of the growing colonies on the L.B. acetate agar. This was indicated by the researchers36, which explained the bacterial role in calcite formation due to its metabolic activity that increases the pH and deposition of calcite on its surface (figure 6).
CONCLUSION
Bacterial species differ according to the environments in which they live, as the species that are isolated from clinical sources and possess many virulence factors that make them more dangerous and pathogenic to humans differ about the same species if separated from a variety of external environments, which makes them virulent or have new characteristics that make them adapted to live in the domains from which they are isolated
Funding: "This research received no external funding,"
Acknowledgments: The author would like to thank Biology depart. , University of Mosul for supporting this study.
Conflicts of Interest: "The authors declare no conflict of interest."
1. Halford B., Mighty microbes lurking in the kitchen, Extreme environments: Dishwasher edition,2018, Volume 96, Issue 12.
2. Malik, A.; Grohmann, E.; Alves, M. Management of Microbial Resources in the Environment, 1st ed.; Springer: Amsterdam, The Netherlands, 2013. [CrossRef]
3. Patel, K.; Amaresan, N. Antimicrobial Compounds from Extreme Environment Rhizosphere Organisms for Plant Growth. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 651–664.
4. Yong, Y.C.; Zhong, J.J. Recent Advances in Biodegradation in China: New Microorganisms and Pathways, Biodegradation Engineering, and Bioenergy from Pollutant Biodegradation. Process Biochem. 2010, 45, 1937–1943. [CrossRef]
5. Sikkema R., Koopmans M. One Health training and research activities in Western Europe. Infection Ecology and Epidemiology. 2016; (6):1-9. doi:10.3402 / iee.v6.33703.
6. World Organization for Animal Health (OIE). Animal Production Food Safety. 2016.: http://www.oie.int/en/foodsafe...
7. Cray J.A., Bell A.N.W., Bhaganna P., Mswaka A.Y. et al. The biology of habitat dominance; can microbes behave as weeds?. Microb. Biotechnol. 2013; (6):453492.
8. Williams, P., and J. M. Tomas. The pathogenicity of Klebsiella pneumoniae. Rev Med Microbiol 1990;1:196–204.
9. Montgomerie, J. Z. Epidemiology of Klebsiella and hospitalassociated infections. Rev Infect Dis 1979;1:736–53.
10. Podschun, R., and U. Ullmann. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev 1998;11:589–603.
11. Kaul S, Brahmadathan KN, Jagananati M, Sudarsanam TD, Pitchamuthu K, Abraham OC, John G. One Year Trends in the Gram Negative Bacteria antibiotic susceptibility patterns in a Medical Intensive care unit in South India. Indian J Med Microbiol 2007;25:230-5.
12. Podschun R, Pietsch S, Holler C, Ullmann U. Incidence of Klebsiella species in surface waters and their expression of virulence factors. Appl Environ Microbio 2001;67(7):3325- 3327.
13. Russell, F. M., Biribo, S. S. N., Selvaraj, G., Oppedisano, F., Warren, S., Seduadua, A.,... & Carapetis, J. R.. As a bacterial culture medium, citrated sheep blood agar is a practical alternative to citrated human blood agar in laboratories of developing countries. Journal of clinical microbiology,2006; 44(9), 3346-3351.
14. Esselmann TM, Liu PV. Lecithinase production by Gram negative bacteria. J Bacteriol 1961;81(6):939-45.
15. Cappuccino JG, Sherman N. Microbiology - A laboratory manual 1996;159-201.
16. Panus. E, Chifiriuc M. B., Bucur M. Virulence, pathogenicity, antibiotic resistance and plasmid profile of Escherichia coli strains isolated from drinking and recreational waters, in 17th European Congress of Clinical Microbiology and Infectious Diseases and 25th International Congress of Chemotherapy, 2008.
17. Hassan A,Usman J, Kaleem F,et al. Evalution of different detection methods of biofilm formation in the clinical isolates.Braz J Infect Dis,2011; 15(4) 305-311.
18. Lee NY. Calcite Production by Bacillus amyloliquefaciens CMB01, J. Microbiol., 2003; 41(4): 345-348.
19. Hamilton,W.R. ; Woolley,A. R. and Bishop, A. C..Minerals Rocks and Fossils.The Hamlyn Publishing Group Limited,1986; Italy.
20. Babič, M. N., Gostinčar, C., & Gunde-Cimerman, N.. Microorganisms populating the water-related indoor biome. Applied Microbiology and Biotechnology, 2020;104(15), 6443-6462
21. Weikl F, Tischer C, Probst AJ, Heinrich J, Markevych I, Jochner S, Pritsch K. Fungal and bacterial communities in indoor dust follow different environmental determinants. PLoS One.2016; 11: e0154131. https://doi.org/10.1371/journal.pone.0154131.
22. Jayaprakash B, Adams RI, Kirjavainen P, Karvonen A, Vepsäläinen A, Valkonen M, Järvi K, Sulyok M, Pekkanen J, Hyvärinen A, Täubel M. Indoor microbiota in severely moisture damaged homes and the impact of interventions. Microbiome.2017; 5(1):138. https://doi. org/10.1186/s40168-017-0356-5.
23. Shan Y, Wu W, Fan W, Haahtela T, Zhang G . House dust microbiome and human health risks. Int Microbiol.2019; 22(3):297–304. https://doi.org/10.1007/s10123-019-00057-5.
24. Pereira, S. C. L., & Vanetti, M. C. D. . Potential virulence of Klebsiella sp. isolates from enteral diets. Brazilian Journal of Medical and Biological Research,2015; 48, 782-789.
25. Albesa, I. Klebsiella pneumoniae haemolysin adsorption to red blood cells. Journal of applied bacteriology,1989; 67(3), 263-266.
26. Gundogan N, Citak S, Yalcin E. Virulence properties of extended spectrum beta-lactamase-producing Klebsiella species in meat samples. J Food Prot 2011; 74: 559-564, doi: 10.4315/0362-028X.JFP-10-315.
27. Kalaivani et al.,. Clinical isolates of Klebsiella pneumoniae and its virulence factors from a tertiary care hospital. Original Research Article,2019;023.
28. Singh, B. R., Sharma, V. D., & Chandra, R.. Detection, prevalence, purification and characterization of lecithinase of Klebsiella pnemoniae.1999.
29. Ali, M. J., & Al-Rikabi, R. H. December). Antibiotic and virulence profile of UTIs associated bacteria. In AIP Conference Proceedings.2020; (Vol. 2290, No. 1, p. 020008). AIP Publishing LLC. 15.
30. Yin, W., Wang, Y., Liu, L., & He, J. Biofilms: the microbial “protective clothing” in extreme environments. International journal of molecular sciences, 2019;20(14), 3423.
31. Seifi, K., Kazemian, H., Heidari, H., Rezagholizadeh, F., Saee, Y., Shirvani, F., & Houri, H. Evaluation of biofilm formation among Klebsiella pneumoniae isolates and molecular characterization by ERIC-PCR. Jundishapur journal of microbiology, 2016;9(1).
32. Aljanaby, A. A. J., & Alhasani, A. H. A.. Virulence factors and antibiotic susceptibility patterns of multidrug resistance Klebsiella pneumoniae isolated from different clinical infections. African Journal of Microbiology Research, 2016;10(22), 829-843.
33. Abbas, O. N., Mhawesh, A. A., & Al-Shaibani, A. B. Molecular Identification of Pathogenic Klebsiella pneumoniae Strains Producing Biofilm. Medico Legal Update,2020; 20(3), 1068-1074.
34. Bosak T. Calcite precipitation, microbially induced. In: Reitner J, Thiel V (eds) Encyclopedia of earth sciences series. Springer, Netherlands, 2011;pp 223–227.
35. De Muynck W, De Belie N, Verstraete W. Microbial carbonate precipitation in construction materials: a review. Ecol Eng.2010; 36:118–136.
36. Chahal, N., Rajor, A., & Siddique, R. Calcium carbonate precipitation by different bacterial strains. African Journal of Biotechnology, 2011;10(42), 8359-
Received: 2 January 2023/ Accepted: 19 April 2023 / Published:15 June 2023
Citation: Matter I R, AL-Omari A W, Almola A H. 16SrRNA sequencing analysis for identification of Klebsiella pneumoniae isolated from the extreme kitchen environment. Revis Bionatura 2023;8 (2) 24. http://dx.doi.org/10.21931/RB/2023.08.02.24
