2022.07.01.31
Files > Volume 7 > Vol 7 No 1 2022
Study single nucleotide polymorphism in Promoter region of UGT1A1 Gene in Iraqi Patients with Gilbert's syndrome
1* Department of Biology, Al-Rasheed University College, Baghdad, Iraq;
2 Al-Rafidain University College, Baghdad, Iraq
3 Biology department Wasit University College of science. Iraq
Available from: http://dx.doi.org/10.21931/RB/2022.07.01.31
ABSTRACT
This study aimed to detect genetic variants of the UGT1A1 gene in patients with Gilbert's syndrome. To detect this, primers were designed; PCR and direct sequencing were done for the promoter area of the gene as a diagnostic tool for the detection of any polymorphism. Variation and polymorphism were detected within the promoter mutants of the UDP glycosyltransferase _UGT1A1 gene that causes hyperbilirubinemia in a group of Iraqi patients compared with a group of the normal healthy individual as controls. The patients with hyperbilirubinemia in this study were 30 in which the total bilirubin level was more than 12 mg/dl serum; they included 25 males and 5 females, while the control group consisted of 20 healthy individuals. This study was carried out from September 2019 till April 2021. The result displayed high occurrence of Gilbert syndrome within male patients than in females, and regarding the analyses of mutation of bilirubin UDP glycosyltransferase _UGT1A1 gene, it is clear that the genotypic distribution of variation among the hyperbilirubinemia patients included all 30 patients, while SNP was detected in 18 patients out of 30 which indicate that the UGT1A1 gene mutation was a likely risk factor for the development of hyperbilirubinemia related Gilbert syndrome in Iraq. The homozygous and heterozygous polymorphisms A/G inside the promoter region of the UGT1A1 gene were effectively identified by sequencing. Our finding suggests that TA repeats and allele of UGT1A1 polymorphism A/G are associated with Gilbert's syndrome and act as genetic markers of this disease in Iraqi patients. To analyze data and sequence variation in gene, generous software was used after amplifying the gene. All processes include DNA extraction, PCR amplification, sequencing, and assembly.
Keywords. Gilbert's syndrome, UGT1A1 gene, polymorphism, hyperbilirubinemia
INTRODUCTION
Gilbert syndrome (GS) is a disorder conspicuous by intermittent unconjugated hyperbilirubinemia and jaundice1 Protein formed from the UGT1A1 gene, named the bilirubin uridine diphosphate glucuronosyltransferase (bilirubin-UGT) enzyme, is a hepatic enzyme in which glucuronidated bilirubin; a material formed once red blood cells are broken down. The enzyme's function changes the toxic form of bilirubin to its nontoxic form, making it capable of being thawed and detached from the body; this enzyme is required for the conversion (conjugation) and subsequent elimination of bilirubin from the body,2,3. Gilbert's syndrome is supposed in patients that have unconjugated hyperbilirubinemia produced by reduced activity of the UDP-glucuronosyltransferase 1A1 (UGT1A1) gene in the lack of irregular liver function and hemolysis, noticeable by unbalanced unconjugated hyperbilirubinemia, typically due to the polymorphism uridine diphosphate-glucuronosyltransferase inherited defects in uridine diphosphogluconurate glucuronosyl-transferase (UGT) that encoded by the UGT1A1 gene at chromosome 2q37.1, the reduced enzyme action are lead to hyperbilirubinemia. The variation in the UGT1A1 gene can reason for Gilbert syndrome that characterized by phases of insignificant unconjugated hyperbilirubinemia, 3 The main genetic variants in Gilbert's syndrome are TATA-box repeats of the promoter region that are responsible for the manufacturing of the bilirubin-UGT enzyme and exon 1 G211A of the coding region, UGT1A1 gene mutations either decrease the affinity of UGT1A1 toward bilirubin or decrease enzyme activity. New studies presented serum bilirubin is related to a genetic variation of the UGT1A1 locus 4. In several people, the maximum shared change cause Gilbert syndrome happens in part near the UGT1A1 gene called the promoter region. This alteration must happen in both copies of the UGT1A1 gene to cause Gilbert syndrome. 5, 6,7,8,9
Homozygous polymorphism is the most public UGT1A1 genotype responsible for Gilbert's syndrome when A(TA)7 TAA in the gene's promoter region; this will make a 70-80% reduction in the gene's promoter region glucuronidation activity. In contrast, the change at nucleotide 211 (G211A), which change arginine to replace with glycine at position 71 in the coding region of the UGT1A1 gene, is accountable for around 20% of Gilbert's syndrome cases in Asian people10,11,12Therefore this study was aimed to detect the gene variation in UGT1A1 gene in blood samples as a diagnostic tool by PCR and sequencing in patients from Iraq and determine its susceptibility to with Gilbert's syndrome.
MATERIALS AND METHODS
This study was held from September 2019 to April 2021 at the University of Al-Rasheed / Biology Department, and the molecular study was held in the molecular laboratory of ASCo. They were a learning center in Iraq. Blood samples were collected from a population consisting of 30 subjects with Gilbert syndrome disease who complained of high bilirubin level in blood and have familial history of Gilbert syndrome with no clinical features refer to a disorder of liver functions; they were selected from those attending private Hospital in Baghdad. The age of these patients ranged from 17 - 55 years, in which 25 patients were males, and only 5 were female. The control group involved 20 healthy individuals were their ages and sex thoroughly similar to the patients and had no history of any disease.
An aliquot of 5 ml blood sample was gained from the vein of patient and control groups and divided into two parts, the first part (2 ml) was collected into an EDTA tube and kept at -20 °C for molecular analyses while serum was used for biochemical tests for liver enzyme ALT, AST, alkaline phosphatase, was recovered from the second blood part (3 ml).
Biochemical analyses of liver enzymes ( ALT, AST, alkaline phosphatase ) were estimated. The levels of all enzymes were examined for both the experimental and control groups.
DNA extraction
First DNA was isolated according to the procedure ReliaPrep™ Blood gDNA Miniprep System, Promega as the steps in manufacturer producer (table 1)

Table 1. Kits for DNA extraction
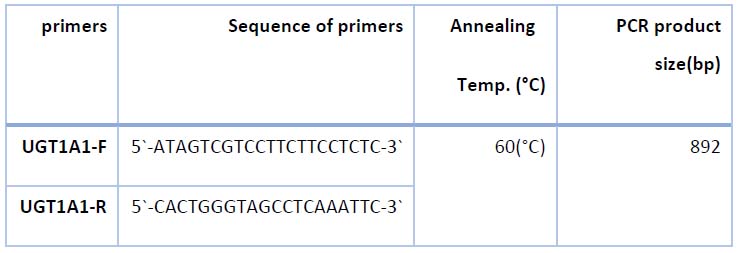
Table 2. sequences of primers used in the detection promoter region of UGT1A1 gene in this study
Polymerase Chain Reaction (PCR)
PCR was performed for 50 samples (30 patients and 20 control groups) using specific primers designed to amplify and sequence the promoter region of the UGT1A1 gene as described in table (2). PCR amplifications were performed as presented in (table 3)
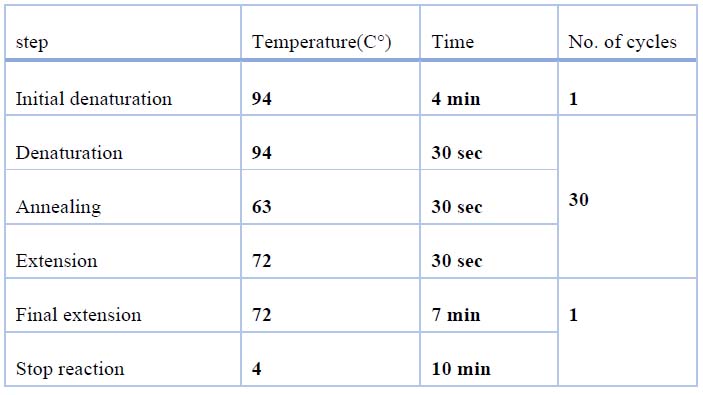
Table 3. PCR amplification program of UGT1A1 gene
DNA Standard Sequencing
To analyze the nucleotides sequences for all samples to determine the presence of single nucleotide polymorphism (SNP) and determine the genetic variation of this gene within the population, sequence analysis of the promoter area of the UGT1A1 gene was done accomplished on Gilbert syndrome patients. PCR product was directed for Sanger sequencing by ABI3730XL, automated DNA sequences, Macrogen Corporation – Korea. The obtained results were established through email and investigated by generous software.
RESULTS
Liver function test results of Gilbert syndrome patients
The patients with hyperbilirubinemia suffering from Gilbert syndrome in this study were 30 by a whole bilirubin level of more than 12 mg/dl, which comprise 25 patients were males and 5 patients were females, samples for patients and control groups have normal liver enzyme ALT, AST, alkaline phosphatase values.
Distribution of Cases According to Gender
the distribution of GS according to gender showed higher rates in males than females when the male cases were 83.33% and 16.66% for females.(figure 1)
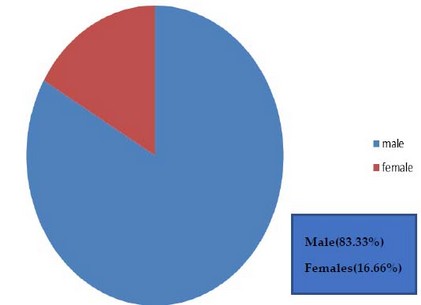
Figure 1. Distribution of Cases According to Gender
Purity and concentration of DNA extracted from
The DNA was effectively extracted from blood samples. The purity of DNA extracted from blood samples was extended from 1.8 to 2, and the concentration of DNA was extended from 70 -120 µg/ml.
Amplification of UGT1A1 gene
The molecular genetic markers related to patients with hyperbilirubinemia were examined, the segment of the UGT1A1 gene can be amplified by using the PCR, including the promoter region with a size of about 892bp shown in figure (2). The primers in this study were designed using the NCBI Primer-Design online tool to detect single nucleotide polymorphism (SNPs)that may be found in the target gene leading to hyperexpression bilirubin that leads to Gilbert syndrome.
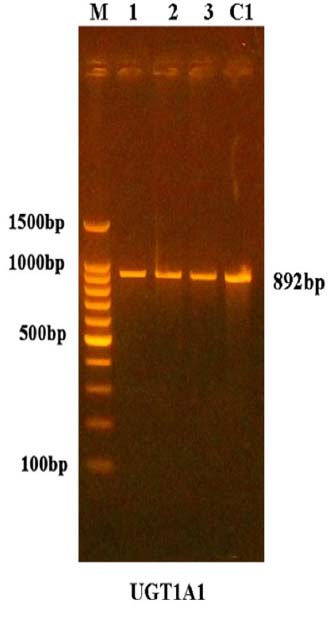
Figure 2. The amplification of UGT1A1 gene promoter region of patient M= 100bp ladder marker. Lanes 1-C1 is 892bp PCR products.
Sequencing and Analysis of UGT1A1 gene
An investigation of any genetic variation and mutations in the UGT1A1 gene was done. The sequence analysis result for the promoter region in the UGT1A1 gene showed there were two genetic variants within the gene's promoter area. The first variant was, 8 repeats of TA copies was observed in the sequence of the gene of all patients samples; the results were directly compared with the Iraqi healthy control group in which it's clear that 7 TA repeats were detected in this region as shown in table 4; also it is compared databases at www.ncbi.nlm.nih.gov by the BLAST check-up tool and besides using Genus software program figure (3 –a,b). Also, there is the second variant in the promoter region of UGT1A1 region of the gene was detected in which among the thirty Gilbert Syndrome patients, there are one SNP detected in the promoter region, which is A substitution with G in site -208, as shown in table 4, and the number of patients who had this polymorphism (A instead of G) was 18 out of 30 patients (60 %) in comparison with that of healthy individuals included in this study, while the other 12 patients (40%) no variation appeared in this studied part of the gene, might have in another coding region of UGT1A1 gene, as shown in figure (4-a,b)

Table 4. Genetic variation detected in the promoter area of UGT1A1 gene
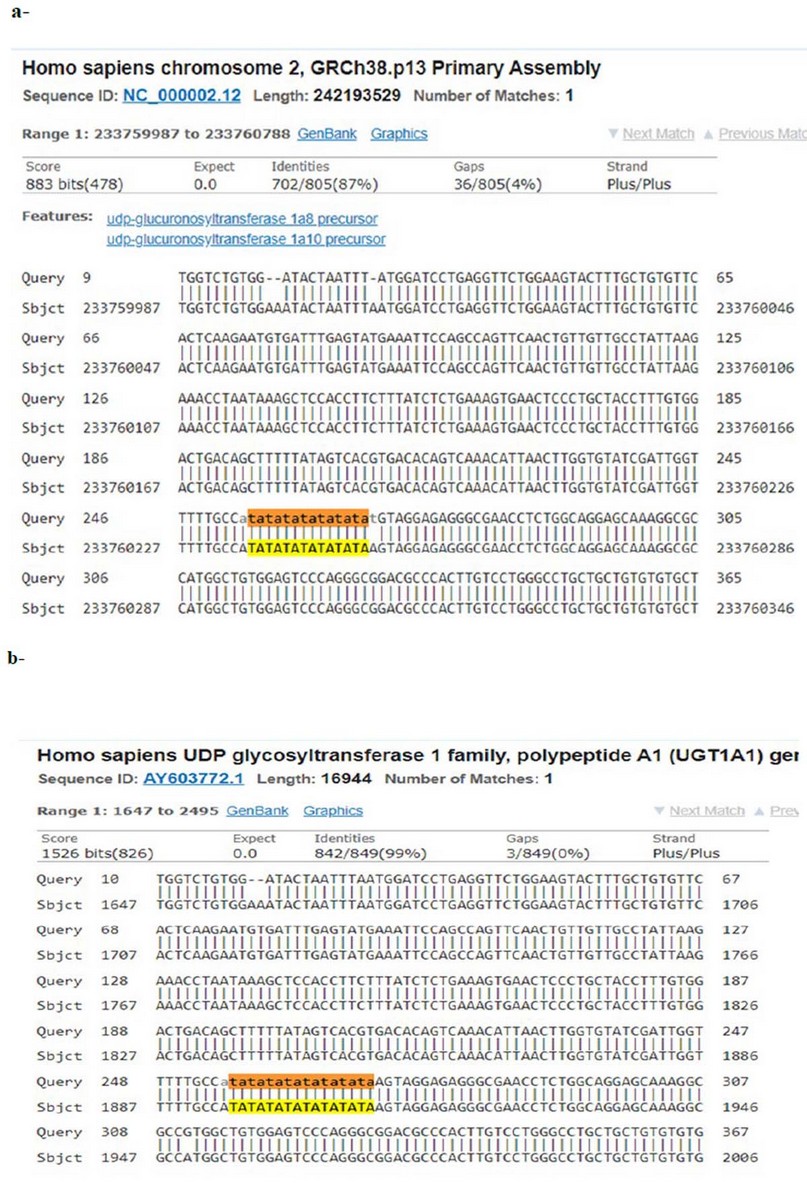
Figure 3. A-Alignment demonstrates promoter area of UGT1A1 gene a- control, B- Gilbert syndrome using a sequencer analyzed by BLAST tool. Query number represented the present data, whereas the subject represented the reference gene sequence
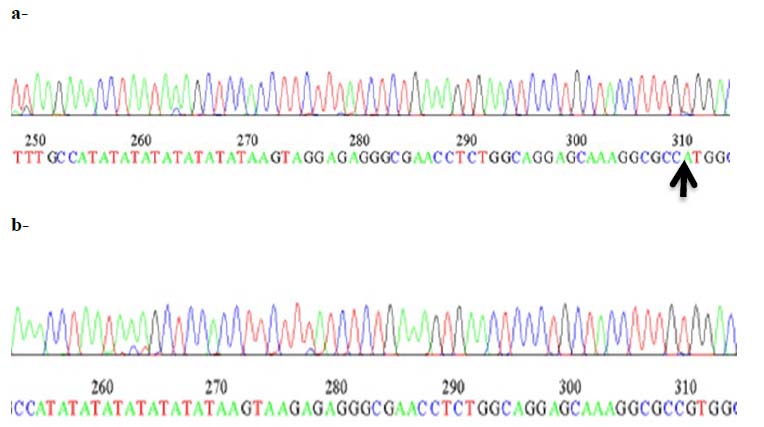
Figure 4-. a,b Nucleotide sequencing profile of promoter region in UGT1A1 gene polymorphism at position -208 as recorded by ABI3730XL, automated DNA sequences.
The outcomes of PCR analysis of UGT1A1 (-208 A/G) polymorphism subdivided GS patients into three genotypes groups: the first group is homozygous wild type (AA) in which no variation appeared when they have two identical alleles for a gene, the second group is homozygous mutant (GG), and the third group is heterozygous (AG) having two different alleles for a gene one dominant and one recessive, this is a heterozygous mutation, As a result of this observation, AG genotype may be considered as a risk factor, while GG genotype appeared only in 8 patients out of 30 patients (26.6%) when they harbored two AA/GG variations and cannot be considered as a protective factor for Gilbert syndrome in the Iraqi population examined. This observation indicated that these genotypes are associated with Gilbert's genetic predisposition. The percentage of genetic variation of the prompter region of UGT1A1 (-208 A/G) was illustrated in Table 5.

Table 5. Genetic variation in the promoter region of UGT1A1 gene of Gilbert syndrome patients
DISCUSSION
In this study, the distribution of patients with GS according to gender showed higher rates of infection in males (25 out of 30) (83.33% ) than females (5 out of 30)(16.6%), when this results supported by the results of, Sieg A. et al. (1987). who found that males are more frequently affected than females, this might be clarified by the existence of a high bilirubin load per kilogram body weight in males, also may the androgen steroid inhibition of bilirubin enzymatic glucuronidation, Bosma PJ, et al. (1995) 20,21
The promoter area of the UGT1A1 gene in Gilbert syndrome patients compared to control samples(healthy individuals) showed two types of variation; this is agreed with Canu. et al. 2013, showed that more than 130 variations in both regulatory and coding regions of UGT1A1 had been recognized in hereditary hyperbilirubinemia patients 22. the first one was the repeats of (TA) copies in promoter region appeared in all patients in which it repeated 8 times in sequences of patients while 7 times repeated in the sequence of the control group,
Bosma et al., 1995; Monaghan et al., 1996; Beutler et al., 1998; Biondi et al., 1999; Kadakol et al., 2000; Farheen et al., 2006), 14,15,16,17,18,19, who found that the determined mutual UGT1A1 genotype accountable intended for Gilbert's syndrome, in which the homozygous polymorphism A(TA)7 TAA found within promoter area of the gene, make a 70-80% decrease in glucuronidation activity, when they repeated 7 times in a sequence of patients and 6 times in control healthy group, while in our Iraqi patients it repeats 8 times in compared with the sequence of healthy patients, it repeats only 7 times, (rs 3064744).
The second variation in the promoter area of the UGT1A1 gene show one SNP in 18 patients, A substitution with G in -308 site in the promoter region of the gene indicate that these mutations show a part in this disease in which these polymorphisms modify the expression that altered transcription factor gene binding. Genetic factors are considered as an essential factor for the disease, and it's clear that the variation in the promoter area of the gene contributes to GS disease, In which the promoter area controls the manufacture of the bilirubin-UGT enzyme.
Results were the same in the study Xiao-xiao Mi ., 2019, which detected the association between Gilbert syndrome and UGT1A1 G/C-46 SNP (rs873478) in the proximal promoter region gene and Gilbert syndrome.
Besides, homozygous genotypes (AA/GG) in the promoter region were more frequent in the patients than in the controls. These results disagree with another study in which they found this variation in exon 1while in our study, one heterozygous SNPs were detected (AA/AG) in eight Gilbert syndrome samples.
Our finding agreed with Xiao-xiao Mi et al., 2019 13, who screened many coding areas of the UGT1A1 gene in 60 Gilbert syndrome patients, who noted proximal promoter area of UGT1A1 variation in 85% of patients, but no mutations were identified (Glasow et al., 2001). New genome association studies exposed that serum bilirubin is related to a genetic variation of the UGT1A1 locus 9.
Changes in the UGT1A1 gene either decrease the affinity of UGT1A1 toward bilirubin or reduce enzyme activity 10. The patients may have changes in another coding region of the UGT1A1 gene.
Single nucleotide polymorphisms (SNPs) were the most common variation in a specific gene that influences how a person responds to the environmental factor, which may alter the disease risk.23
CONCLUSIONS
In the present study, our results indicated that variation on the promoter region of the UGT1A1 gene affects the bilirubin level; TA copies repeats were 8 times in a sequence of patients while 7 times in control healthy samples.UGT1A1 (-208 A/G) polymorphism was associated with Iraqi patients with Gilbert syndrome. However, it should also be noted that the GG and AG genotype could be considered markers of genetic predisposition to higher levels of bilirubin in the Iraqi population, thereby leading to increased susceptibility to Gilbert syndrome. In conclusion, the UGT1A1 gene mutation was a possible risk factor for Gilbert syndrome in Iraq
Author Contributions:
-Conceptualization, software, investigation, methodology, data duration, funding acquisition
Marwa A. Kubba; validation, formal analysis, resources, visualization, Abeer Ali Marhoon;
Writing—original draft preparation, writing—review and editing,
Supervision, project administration, Rafed Abbas Kadhum
-All authors have read and agreed to the published version of the manuscript."
Acknowledgment: I am sincerely thankful to Al-Nahrain University, College of Biotechnology for providing me with the opportunity to make paper; I am also thankful to Dr. Sahar M. Hussein for guiding us in many stages during our work.
Funding: "This research received no external funding."
Institutional Review Board Statement:
Ethical Clearance: The ethical research committee at scientific research by ethical approval of both environment and health and higher education and scientific research ministries in Iraq CONFLICT OF INTEREST The authors declare that they have no conflict of interest.
FUNDING: self-funding
Informed Consent Statement: "Informed consent was obtained from all subjects involved in the study."
"Written informed consent has been obtained from the patient(s) to publish this paper".
Conflicts of Interest: Nile
REFERENCES
1- Bosma PJ. Inherited disorders of bilirubin metabolism. J Hepatol 2003; 38:107–17
2- Carpenter, S.L., Lieff, S., Howard, T.A., Eggleston, B., & Ware, R.E. (2008). UGT1A1 promoter polymorphisms and the development of hyperbilirubinemia and gallbladder disease in children with sickle cell anemia. Am J Hematol. 83: 800– 803.
3- Kataoka, R., Kimata, A., Yamamoto, K., Hirosawa, N., Ueyama, J., Kondo, T., Okada, R., Kawai, S., Hishida, A., Naito, M., Morita, E., Wakai, K., & Hamajima, N. (2011). Association of UGT1A1 Gly71Arg with urine urobilinogen. Nagoya J Med Sci. 73(1-2):33-40
4- Johnson, A.D., Kavousi, M., Smith, A.V., Chen, M.H., Dehghan, A., Aspelund, T., Lin, J.P., Dujin, C.M., Harris, T.B., Cupples, L.A., Uitterlinden, A.G., Launer, L., Hofman, A., Rivadeneira, F., Sticker, B., Yang, Q., O’Donnell, C.J., Gudnason, V., & Witteman, J.C.(2009). Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet. 18: 2700–2710
5-Gong QH, Cho JW, Huang T, Potter C, Gholami N, Basu NK, Kubota S, Carvalho S, PenningtonMW, Owens IS, Popescu NC. Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics. 2001 Jun;11(4):357-68.
6- King CD, Rios GR, Green MD, Tephly TR. UDP-glucuronosyltransferases. Curr Drug Metab. 2000Sep;1(2):143-61. Review.page 4
7- Long J, Zhang S, Fang X, Luo Y, Liu J. Association of neonatal hyperbilirubinemia with uridinediphosphate-glucuronosyltransferase 1A1 gene polymorphisms: meta-analysis. Pediatr Int. 2011Aug;53(4):530-40. doi: 10.1111/j.1442-200X.2011.03337.x.
8- Maruo Y, Nishizawa K, Sato H, Sawa H, Shimada M. Prolonged unconjugated hyperbilirubinemia associated with breast milk and mutations of the bilirubin uridine diphosphateglucuronosyltransferase gene. Pediatrics. 2000 Nov;106(5):E59.
9- Owens IS, Basu NK, Banerjee R. UDP-glucuronosyltransferases: gene structures of UGT1 andUGT2 families. Methods Enzymol. 2005;400:1-22. Review.
10- Biondi ML, Turri O, Dilillo D, Stival G, et al. (1999). Contribution of the TATA-box genotype (Gilbert syndrome) to serum bilirubin concentrations in the Italian population. Clin. Chem. 45: 897-898.
11- Kadakol A, Ghosh SS, Sappal BS, Sharma G, et al. (2000). Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler-Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum. Mutat. 16: 297-306.
12- Farheen S, Sengupta S, Santra A, Pal S, et al. (2006). Gilbert's syndrome: High frequency of the (TA)7TAA allele in India and its interaction with a novel CAT insertion in promoter of the gene for bilirubin UDP-glucuronosyltransferase 1 gene. World J. Gastroenterol. 12: 2269-2275
13- Xiao-xiao Mi,1 Jian Yan,1 Xiao-jie Ma,2 Ge-li Zhu,2 Yi-dan Gao,2 Wen-jun Yang,3 Xiao-wen Kong,2 Gong-ying Chen,2 Jun-ping Shi , 1,2 and Ling Gong 2., (2019). nalysis of the UGT1A1 Genotype in Hyperbilirubinemia Patients: Differences in Allele Frequency and Distribution. BioMed Research International. Volume 2019, Article ID 6272174, 9 pages
14-Bosma PJ, Chowdhury JR, Bakker C, Gantla S, et al. (1995). The genetic basis of the reduced expression of bilirubin UDPglucronosyltransferase 1 in Gilbert's syndrome. N. Engl
15-Monaghan G, Ryan M, Seddon R, Hume R, et al. (1996). Genetic variation in bilirubin UPD-glucuronosyltransferase gene promoter and Gilbert's syndrome. Lancet 347: 578-581.
16-Beutler E, Gelbart T and Demina A (1998). Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc. Natl. Acad. Sci. U. S. A. 95: 8170-8174.
17-Biondi ML, Turri O, Dilillo D, Stival G, et al. (1999). Contribution of the TATA-box genotype (Gilbert syndrome) to serum bilirubin concentrations in the Italian population. Clin. Chem. 45: 897-898.
18- Kadakol A, Ghosh SS, Sappal BS, Sharma G, et al. (2000). Genetic lesions of bilirubin uridine-diphosphoglucuronate glucuronosyltransferase (UGT1A1) causing Crigler-Najjar and Gilbert syndromes: correlation of genotype to phenotype. Hum. Mutat. 16: 297-306.
19- Farheen S, Sengupta S, Santra A, Pal S, et al. (2006). Gilbert's syndrome: High frequency of the (TA)7 TAA allele in India and its interaction with a novel CAT insertion in promoter of the gene for bilirubin UDP-glucuronosyltransferase 1 gene. World J. Gastroenterol. 12: 2269-2275.
20- Sieg A, Arab L, Schlierf G, Stiehl A, Kommerell B. Prevalence of Gilbert's syndrome in Germany. Dtsch Med Wochenschr 1987.
21- Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP(1995). The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 Gilberts syndrome.N Eng J Med ., 333(18):1171-5.
22- G. Canu, A. Minucci, C. Zuppi, and E. Capoluongo, "Gilbert and crigler najjar syndromes: an update of the UDP-glucuronosyltransferase 1A1 (UGT1A1) gene mutation database," Blood Cells, Molecules, and Diseases, vol. 50, no. 4, pp. 273–280, 2013
23- M. Sahar, Mohammed H. Wali, A. Marwa and H. Bushra, detection of genetic polymorphism in EGFR gene in triple negative breast cancer women from Iraq. Biochem. Cell. Arch. Vol. 20, No. 1, pp. 1921-1925, 2020
Received: 16 October 2021 / Accepted: 20 November 2021 / Published: date. 15 February 2022
Citation: Kubba, M. A.; Marhoon , A. A.; Kadhum , R. A. Study single nucleotide polymorphism in Promoter region of UGT1A1 Gene in Iraqi Patients with Gilbert's syndrome. Revis Bionatura 2022;7(1). 31. http://dx.doi.org/10.21931/RB/2022.07.01.31
