2023.08.03.61
Files > Volume 8 > Vol 8 No 3 2023
Structural analysis and cytotoxic evaluation of kisspeptin10 and analogs in types of cancer
1 Programa de Medicina, Facultad de Ciencias de la Salud, Universidad Autonoma de Bucaramanga; [email protected]
2 Instituto de Investigación Masira, Facultad de Ciencias Médicas y de la Salud, Universidad de Santander; [email protected]
* Correspondence: [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.03.61
ABSTRACT
The Kisspeptin system is a peptidergic system that plays a crucial role in regulating of reproduction and hormonal function. Kisspeptin is a peptide synthesized from the KiSS-1 gene and has been identified as the endogenous ligand of the kisspeptin receptor (KISS1R or GPR54 receptor). This system plays a key role in activating sex hormone secretion and puberty. In addition to its function in the regulation of reproduction, the Kisspeptin system has been found to play a role in other physiological processes, such as the regulation of appetite, energy metabolism, cardiovascular function, and cancer. In this study, several Kisspeptin analogs with structural modifications were designed and synthesized. The Kisspeptin analogs were evaluated by in vitro cytotoxicity tests on cancer cells of different cancer types. Cell viability assays were performed, and the concentrations that inhibited cell growth by a significant percentage were determined. The results showed that certain Kisspeptin analogs exhibited increased selective cytotoxicity in cancer cells compared to healthy cells.
In conclusion, this study demonstrates that structurally modified Kisspeptin analogs have the potential to be therapeutic agents against some types of cancer. Understanding the structure-activity relationship of these analogs and their evaluation of their selective toxicity on cancer cells will be of great importance.
Keywords: Kisspeptins Analogs, GPR54, Cancer, Cytotoxicity, Molecular Docking, Structure-activity relationship, Anticancer therapy, Drug Design.
INTRODUCTION
The Kiss1R receptor, or GPR54, was discovered and cloned from the rat brain in 19991. In humans, it was mapped on chromosome 19p13.3, encoding a protein of 396 amino acids (75 kDa)2,3. It is widely distributed throughout the central nervous system3 High levels of Kiss1R have been observed in the cerebral cortex, cerebellum, thalamus, and medulla3 Peripherally has been detected in the heart, skeletal muscle, kidney, liver, and placenta2-4 Kisspeptins (KPs), the physiological ligand for the Kiss1R receptor was identified by several groups in 20011,3,4. KPs are several structurally related amidated peptides derived from the differential proteolytic processing of a common precursor of 145 amino acids encoded by the KISS1 gene. The Kiss1R activation leads to regulate the gonadotropin-releasing hormone (GnRH) secretion5, leading to the regulation of luteinizing hormone (LH) and follicle-stimulating hormone (FSH) from the pituitary gland6-8.
Emerging evidence suggests that Kisspeptin plays a crucial role in the development of suppression of metastasis. The metastatic suppressor activity of the kisspeptin system was first observed in melanoma9, where the low levels of KissR expression lead to increased metastasis risks. It has been shown that the role of the KP system is not only limited to this type of cancer but has expanded to other cancers as well. Martin et al.,10 conclude that KP may be directly associated with the aggressiveness of breast cancer and may be pro-invasive in breast tumors. In pancreatic cancer, several authors11-13 suggest that the most damaging aspect of the disease can be predicted and, in some cases, prevented by the KP system, as they observed significant differences in the expression of KISS1 and Kiss1R in healthy and pancreatic cancer cells. Concerning ovarian and endometrial cancer, the KP system improves the prognosis of these patients due to its metastasis suppressor activity14-16. Another interesting approach is in gastric cancer, where high blood levels of kp54 in early-stage gastric cancer patients suggest a possibility that kp54 may function as a marker for diagnosis of this disease17, as well as overexpression of the KiSS-1 inhibits cell growth, proliferation, and invasion in gastric carcinoma cells, proving the antimetastatic potential of kisspeptin17-19. Finally, in prostate cancer, KiSS-1 expression levels were significantly higher in benign prostate cells compared with primary and metastatic prostate cancer cells. Therefore, it is speculated that KiSS-1 may be used as an essential prostate cancer marker since it can be used to monitor the transformation of benign disease to malignancy20-22.
This paper will study the structure-activity relationship of Kisspeptin-10 analogs and evaluate their cytotoxicity, specifically in cancer cells. It seeks to understand how structural modifications in Kisspeptin-10 analogs may influence their antitumor activity and their ability to induce cell death in cancer cell lines. Through in vitro cytotoxicity tests, the viability and response of cancer cells to these analogs were evaluated, which will allow the identification of those with the most significant therapeutic potential.
MATERIALS AND METHODS
AlaScan with Molecular Docking
The 3D structure of the KissR receptor was predicted using AlphaFold2 of ColabFold23, and the quality of the best-predicted model was checked with ProSA-web online tool 24. The design of the Kisspeptin peptide was expected with the PEP-FOLD3 online tool 25, and the top model of the best cluster was selected as the initial model. The kisspeptin peptide was located in the outer membrane domain of the KissR receptor, and each residue of the peptide was mutated to Alanine by using the mutagenesis option of the Pymol tool26. In total, 11 molecular docking simulations were performed, i.e., one with the native peptide and one for each mutated residue. The molecular docking simulations were performed with the FlexPepDock server 27, allowing the peptide's flexibility during the simulations. The possible molecular interactions of the top 3 models for each docking simulation were analyzed with Prodigy online tool28 and BIOVIA Discovery Studio Software29.
Synthesis and chemical characterization of peptides
Each KP10 analog (See Table 1.) was manually assembled on 100 mg of Rink resin using the HBTU/HOBt activation protocol for Fmoc solid-phase peptide synthesis30. In summary, the N-terminal protecting group was removed with 20% piperidine. Previously, the amino acids were activated with HOBt/HBTU 0.45M, and couplings were carried out in the presence of DIPEA 1.2M and microwave irradiation. The 1% TNBS test monitored the correct coupling and deprotection. Finally, the peptide was cleaved from the resin with 90%TFA, and Sep Pak C18 reverse phase columns purified peptides. The peptide identification was performed on a Bruker ultra extreme mass spectrometer with 40,000 FWHM resolution, 1 ppm accuracy, 337 nm laser, and a mass range of 500 to 3000 Da in positive mode using SCiLs Lab for MS Imaging software. A supersaturated 2,5-dihydroxybenzoic acid (DHB) solution in ACN/H2O 30:70 with 0.1% TFA was used as a matrix.
Cell Culture
The Universidad Industrial de Santander kindly donated Cervical Adenocarcinoma (HeLa) and Prostate Adenocarcinoma (PC3) cells. HEK293T was provided by Biopetrolabs (ECACC 12022001, Culture Collection). HeLa and HEK293 cells were grown in Dulbecco's Modified Eagle Medium (DMEM – Invitrogen) with 10% fetal bovine serum (FBS – Invitrogen), 100 U/ml penicillin and 100 mg streptomycin at 37ºC in a humidified atmosphere (95% air and 5% CO2). PC3 cells were cultured in Roswell Park Memorial Institute (RPMI) medium with 10% fetal bovine serum (FBS – Invitrogen), 100 U/ml penicillin, and 100 mg streptomycin at 37ºC in a humidified atmosphere (95% air and 5% CO2).
Cytotoxicity MTT assay
In 96-well culture plates (Greiner, Fisher Scientific), 15000 cells/well were seeded in a fully supplemented medium according to the manufacturer protocol (Abcam) and kept at 37°C. After 24 hours, cells were serum starved and then stimulated with 10, 100, 250 and 500 nM KP10 and analogs for 48 hours. MTT solution (100 uL, 0.5 mg/ml) was added to each well, and the plate was incubated at 37°C for 3h, followed by the addition of MTT solvent, then the plate for 15 minutes. The product was quantified by measuring absorbance at 590 nm using a microplate reader RT-2100C from BioTech. The data were analyzed with the GraphPad Prism 8.0 software.
RESULTS
Structure-activity relationship assessment of kisspeptin10 analogs and evaluation of cytotoxicity in cancers is an important research topic in pharmacology and oncology. Structure-activity relationship assessment is an approach used to understand how modifications in the chemical structure of a molecule affect its biological activity. In the case of kisspeptin10 analogs, we seek to determine how this molecule's structure affects its ability to inhibit cancer cell growth.
Evaluating cytotoxicity in cancer types is another crucial aspect of this research. Cytotoxicity refers to the ability of a substance to damage or destroy cells. In the context of cancer, cytotoxicity assessment involves determining whether kisspeptin10 analogs can selectively kill cancer cells without harming healthy cells.
Synthesis and chemical characterization of Kisspeptin 10 analogs.
Eleven kisspeptin10 analogs were initially synthesized, where the amino acid alanine substitution was performed for each amino acid present. This allowed us to perform a screening on the different properties provided by the side chain of the other amino acids and how they can contribute to the interaction with the receptor.
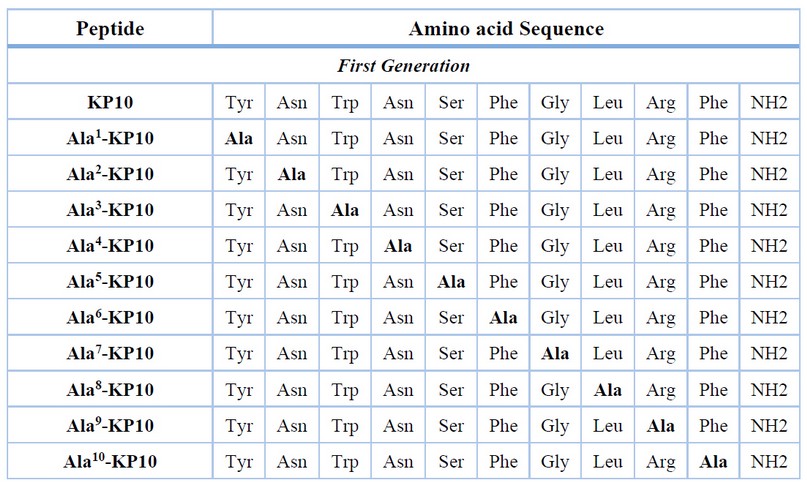
Table 1. Sequences of peptides synthesized and used
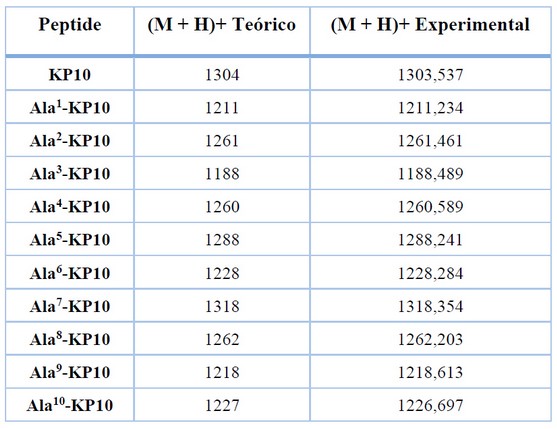
Table 2. Mass spectrometry data. mass-to-charge ratio (m/z) of the [M+H]+ ions of each synthesized peptide.
For all peptides, the molecular weight observed by MS analysis was consistent with the theoretical value (Table 2.)
Evaluation of structure-activity relationships through molecular docking
Evaluating the structure-activity of kisspeptin analogs by molecular docking has proven a powerful tool in pharmacological research. Molecular docking is a computational method that predicts the interaction between a small molecule (ligand) and a macromolecule (receptor) at the molecular level. In the case of kisspeptin analogs, molecular docking is used to predict how these compounds bind and match their target receptor, providing information about their biological activity and potential as therapeutic agents.
Values such as the interface score, which indicates the sum of the energy of the interface residues between kisspeptin analogs and Kiss1R, help select those models with a more extensive interface of interaction, i.e., those models with a more negative value. Moreover, matters such as the predicted binding affinity ΔG, the dissociation constant (Kd), and the percentage of non-interacting surfaces (NIS) charged and apolar also give valuable information about the critical strength between the two molecules in the complexes. Hence, Table 3 shows the values obtained for the top 3 models predicted by the FlexPepDock server. In the case of the interface score, the best deal was acquired by an Ala8-KP10 model (–17.686). The best values were obtained by Ala1-KP10 and Ala3-KP10 models for the predicted binding affinity and the dissociation constant. Finally, the values of the non-interacting surfaces were very similar for all kisspeptin analogs.
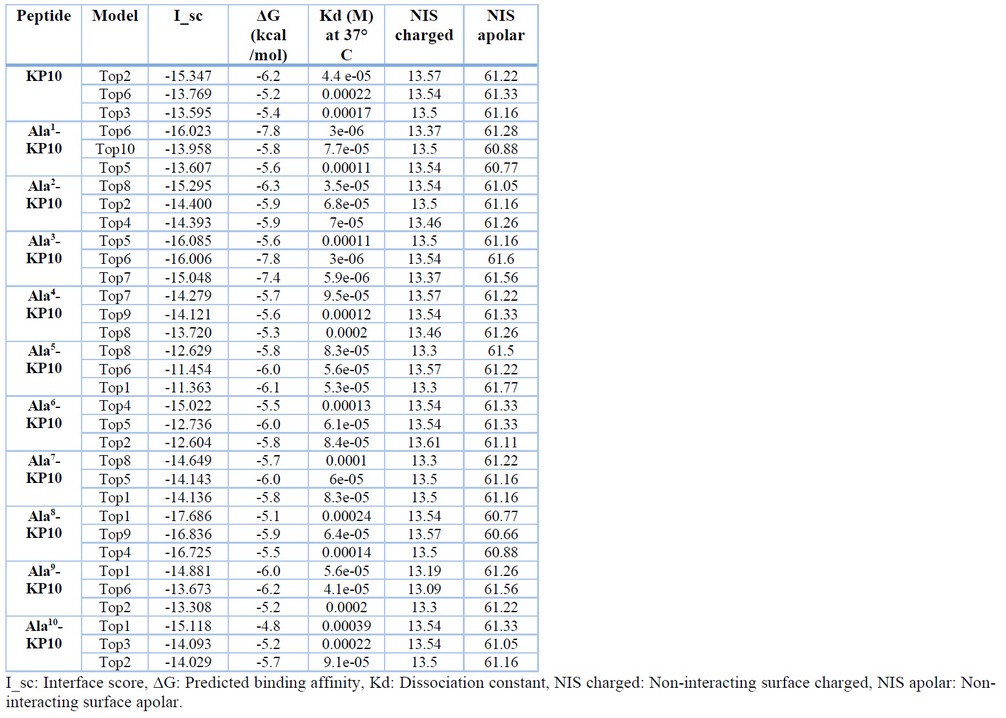
Table 3. Results of the top 3 models according to the interface score (I_sc) predicted by FlexPepDock.
Cytotoxicity Evaluation
All the peptides were subjected to toxicity assay via concentration-response assays on cervix and prostate cancer cell lines. Among these peptides, Ala3-KP10 and Ala4-KP10 demonstrated maximum efficacy against HeLa cells (cervix cancer) with IC50 values of 0.24 ± 0,08 and 0.35 ± 0,08 nM, respectively (as presented in Table 4). Meanwhile, the peptide Ala2-KP10 significantly affected HeLa cells with an IC50 of 3.7 ± 1,25 nM. Conversely, the analogs Ala5-KP10 through Ala10-KP10 only exhibited cytotoxic activity at 500 nM concentrations, limiting their viability under normal physiological conditions (See Figure 1.). The IC50 values for these analogs ranged between 0.27 to 0.41.
In the case of prostate cancer, it can be observed that analogs Ala5-KP10, Ala8-KP10, and Ala10-KP10 achieve high cytotoxicity (IC50 0,16 ± 0,08, 0,14 ± 0,05, 0,05 ± 0,02 respectively). The other analogs show cytotoxicity only at the highest concentration (500 nM). The control of healthy cells was performed in HEK293 (human embryonal kidney) cells, where no traces of cytotoxicity were found with KP10 or any of the tested peptides.
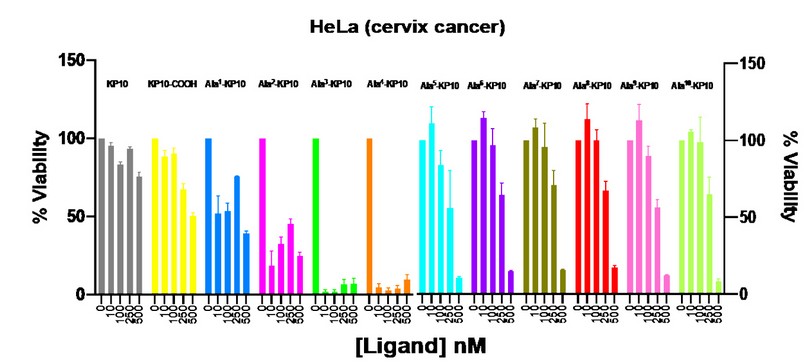
Figure 1. Viability of HeLa cells after stimulation with KP10 and analogs at concentrations of 10, 100, 250 and 500 nM. Viability was measured after 48 hours of incubation with the peptides.
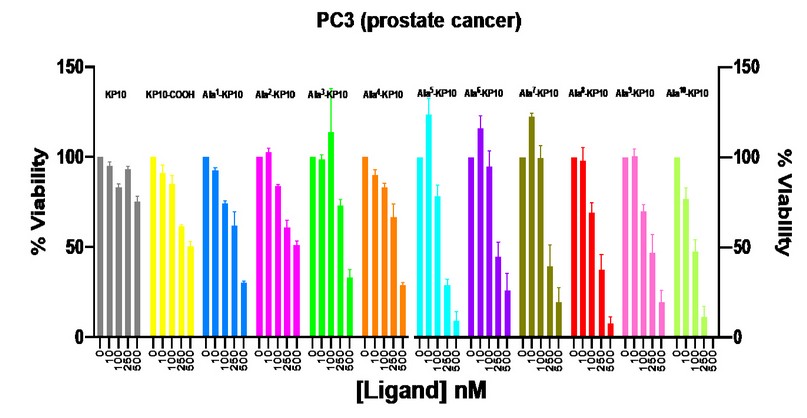
Figure 2. Viability of PC3 cells after stimulation with KP10 and analogs at concentrations of 10, 100, 250 and 500 nM. Viability was measured after 48 hours of incubation with the peptides.
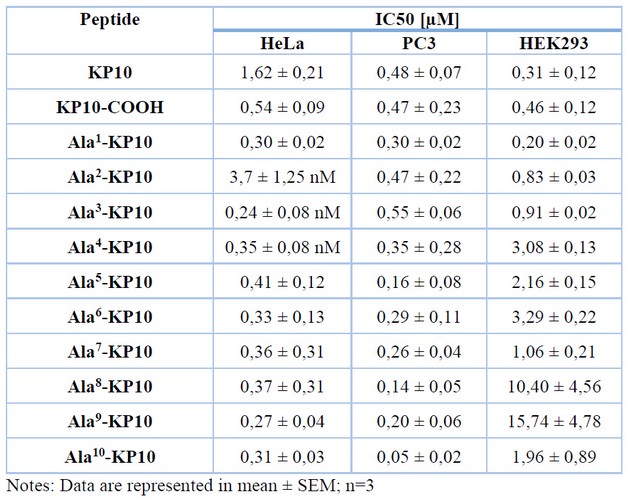
Table 4. The IC50 values of KP10 and analogs on human cancer cell lines and a healthy cell line.
DISCUSSION
Cytotoxicity assays and molecular docking analyses of a series of kisspeptin analogs were performed to evaluate their potential as anticancer therapeutics. The results of the cytotoxicity assays indicate that several analogs have a significant effect on tumor cells, reducing their viability to a variable degree. In cervical cancer, it can be observed that analogs Ala2-KP10, Ala3-KP10, and Ala4-KP10 showed a higher cytotoxic response, which leads to an essential result in cancer research. Evidence indicates that Kisspeptin can increase cell proliferation and invasion of cervical cancer cells31. In addition, Kisspeptin and its receptor are overexpressed in cervical cancer cells31. This suggests that analogs Ala2-KP10, Ala3-KP10, and Ala4-KP10 may behave antagonistically at the Kiss1R receptor expressed on cervical cancer cells. In the docking analyses, analogs Ala3-KP10 and Ala8-KP10 show a higher generation of hydrogen bonds than native Kisspeptin. Figure 4 and the following table illustrate the interactions observed between the respective analogs and the KissR receptor for the best model.
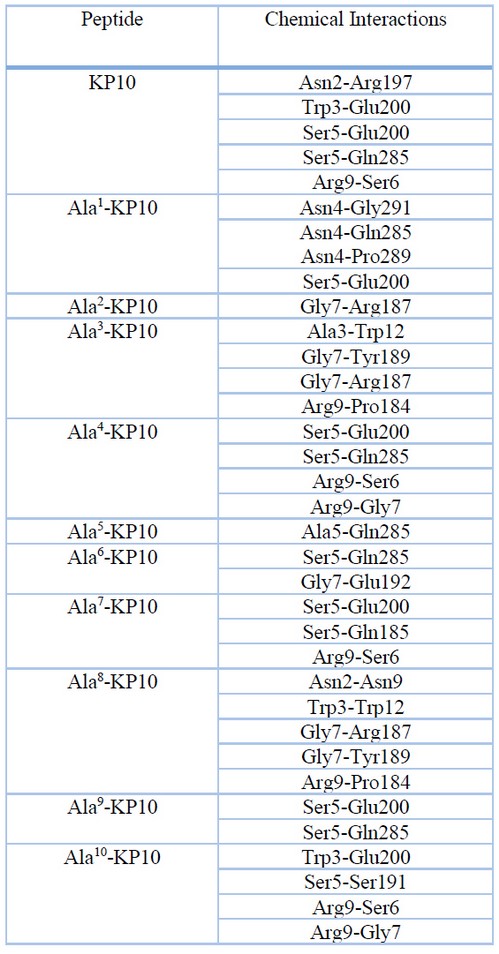
Table 5. Chemical interactions between kisspeptin analogs and the Kiss1R receptor in the best model
A relevant direct Ala3-Trp12 interaction is observed in Ala3-KP10, which allows a possible formation of hydrogen bonds. Tryptophan molecules contain functional groups such as indole and amino groups, which can form hydrogen bonds with the carboxyl group of Alanine. This interaction can stabilize the three-dimensional structure of the proteins in which they are found. In addition, Alanine and tryptophan can interact through dispersion forces, which are weak interactions between non-polar atoms. These forces can contribute to the stability of the protein structure. It is important to note that the interaction between Alanine and tryptophan can vary depending on their position in the amino acid sequence of a protein and the environment in which they are found. The unique properties of each amino acid can influence how they interact.
In Ala8-KP10, when it changed a leucine by Alanine, it is observed that the glycine in position 7 leads in the non-covalent interactions with the receptor. By changing Leu8 to Ala8, two relevant interactions between Gly7 with Arg187 and Tyr189 occur. The new peptide conformation allows a better interaction between these amino acids. As for molecular docking analysis, several significant interactions between kisspeptin analogs and cell proliferation-associated proteins have been identified. In particular, it has been found that the analogs with the highest affinity for these proteins also have the most significant cytotoxic effect.
Other interesting interactions observed with KP10 and analogs and Kiss1R: Interaction between Asn2 of KP10 and Arg197: This interaction involves the formation of a hydrogen bond between the amino group of asparagine (Asn2) in the KP10 peptide and the carboxyl group of arginine (Arg197). This interaction may be necessary for stabilizing the peptide structure and binding to the Kiss1R receptor. Interaction between Trp3 of Ala8-KP10 and Trp12: Here, the tryptophan (Trp3) of KP10 interacts with glutamic acid (Trp12) via hydrogen bonds and possible hydrophobic interactions. These interactions may be crucial for recognition and binding to the Kiss1R receptor. Interestingly, native Kisspeptin also contains a tryptophan at position 3, suggesting a conservation of this interaction in KP10 analogs. Interaction between Ser5 of Ala4-KP10 and Glu200: The serine (Ser5) of Ala4-KP10 forms hydrogen bonds with glutamic acid (Glu200). These interactions may be important in stabilizing the peptide conformation and promoting receptor binding. In native Kisspeptin, the serine is also found at position 5, indicating the conservation of this interaction in KP10 analogs. Interaction between Arg9 of KP10 and Ser6: In this case, the arginine residue (Arg9) of KP10 interacts with the adjacent serine (Ser6). This interaction may play a role in stabilizing the peptide structure and binding to the Kiss1R receptor. This interaction in KP10 analogs suggests a conservation of the interaction between Arg9 and nearby residues in native Kisspeptin.
These differences in ligand-receptor chemical interactions seen by molecular docking simulations do not allow us to deduce the behavior observed in cytotoxicity assays. High cytotoxicity can be observed in cervical cancer cells, medium cytotoxicity in prostate cancer cells, and none in healthy cells (Figure 3). We allude to these different responses between cervical and prostate cancer to the different cellular membranes each of these cells has; we believe that each cell's lipid composition and membrane components make the receptor act differently in each type of cancer studied.
Discussion surrounding these results should focus on how these findings can be used to further the development of kisspeptin analogs as an anticancer therapy. There is a need to examine further the molecular interactions identified and to determine which specific features of the analogs result in the observed cytotoxicity. It is also essential to consider the possibility that these analogs may affect normal cells in the body and how this might be mitigated in clinical use. In addition, it is essential to perform studies on the lipid composition of the membranes of different cells in different types of cancer. It may be relevant to observe the different responses that the kisspeptin system can provide.
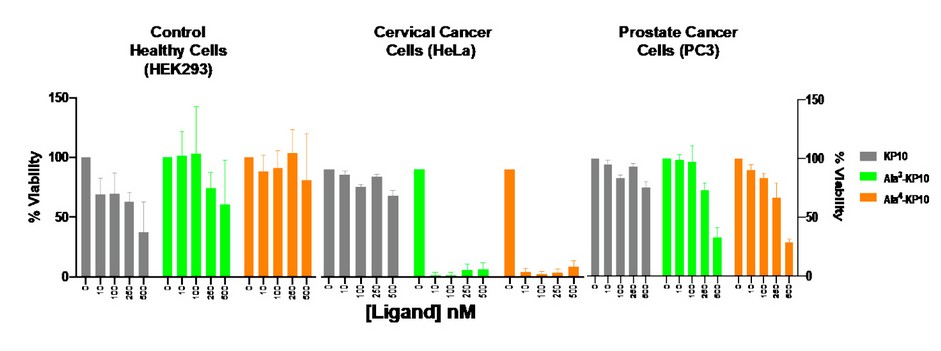
Figure 3. Comparison of analogs Ala3-KP10 and Ala4-KP10 concerning cytotoxicity between healthy cells (HEK293) and cervical (HeLa) and prostate (PC3) cancer cells.
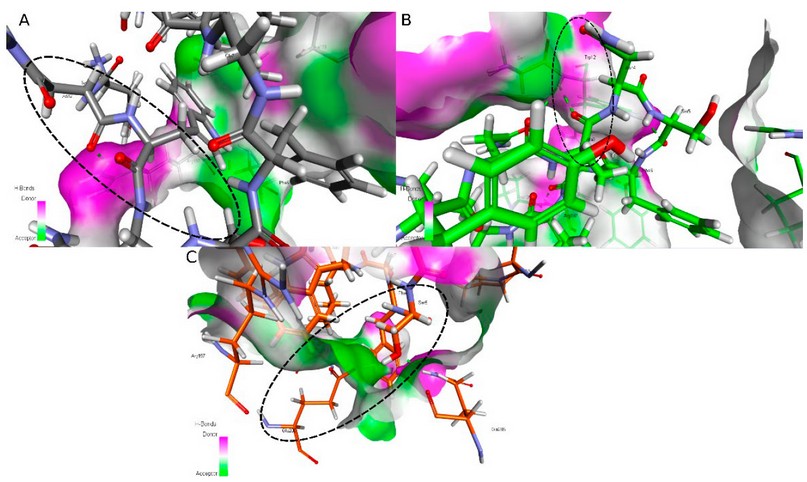
Figure 4. Interesting interactions between KissR receptor and A) KP10, B) Ala3-KP10 and C) Ala4-KP10.
CONCLUSIONS
This study investigated the interactions between kisspeptin10 analogs and the Kiss1R receptor and the cytotoxic effects on cervical and prostate cancer cells. Our results demonstrate that kisspeptin10 analogs can interact successfully with the Kiss1R receptor, suggesting their potential as therapeutic tools in treating these cancers. Furthermore, cytotoxicity assays revealed that kisspeptin10 analogs exhibited significant cytotoxic activity against cervical and prostate cancer cells. These findings further support the therapeutic potential of kisspeptin10 analogs and suggest their possible application in targeted cancer therapy. Importantly, further studies are required to fully understand these interactions' underlying mechanisms and evaluate the efficacy and safety of kisspeptin10 analogs in in vivo models and clinical trials. Nevertheless, our promising results suggest that kisspeptin10 analogs represent an exciting direction in searching for new cervical cancer therapies.
In conclusion, this study provides initial evidence of the interactions between kisspeptin10 analogs and the Kiss1R receptor and their cytotoxic activity in cervical and prostate cancer cells. These findings support the need for future research to fully explore the therapeutic potential of kisspeptin10 analogs in treating these cancers, providing renewed hope for improving patients' quality of life.
Author Contributions: A short paragraph specifying their individual contributions must be provided for research articles with several authors. The following statements should be used “Conceptualization, D.Y.R.S.; methodology, D.J.T.S.; software, P.R.V.; investigation, D.Y.R.S.; writing—original draft preparation, D.Y.R.S.; writing—review and editing, D.Y.R.S, P.R.V.; project administration, D.Y.R.S.; funding acquisition, D.Y.R.S.
Funding: This research was funded by MINCIENCIAS, Government of Colombia. Grant 808-2017.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
1 Lee, D. K. et al. Discovery of a receptor related to the galanin receptors. FEBS Lett 446, 103-107, doi:10.1016/s0014-5793(99)00009-5 (1999).
2 Kotani, M. et al. The metastasis suppressor gene KiSS-1 encodes kisspeptins, the natural ligands of the orphan G protein-coupled receptor GPR54. J Biol Chem 276, 34631-34636, doi:10.1074/jbc.M104847200 (2001).
3 Muir, A. I. et al. AXOR12, a novel human G protein-coupled receptor, activated by the peptide KiSS-1. J Biol Chem 276, 28969-28975, doi:10.1074/jbc.M102743200 (2001).
4 Ohtaki, T. et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature 411, 613-617, doi:10.1038/35079135 (2001).
5 Dhillo, W. S. et al. Kisspeptin-54 stimulates the hypothalamic-pituitary gonadal axis in human males. J Clin Endocrinol Metab 90, 6609-6615, doi:10.1210/jc.2005-1468 (2005).
6 Dungan, H. M. et al. The role of kisspeptin-GPR54 signaling in the tonic regulation and surge release of gonadotropin-releasing hormone/luteinizing hormone. J Neurosci 27, 12088-12095, doi:10.1523/JNEUROSCI.2748-07.2007 (2007).
7 Lapatto, R. et al. Kiss1-/- mice exhibit more variable hypogonadism than Gpr54-/- mice. Endocrinology 148, 4927-4936, doi:10.1210/en.2007-0078 (2007).
8 Seminara, S. B. et al. The GPR54 gene as a regulator of puberty. N Engl J Med 349, 1614-1627, doi:10.1056/NEJMoa035322 (2003).
9 Lee, J. H. & Welch, D. R. Suppression of metastasis in human breast carcinoma MDA-MB-435 cells after transfection with the metastasis suppressor gene, KiSS-1. Cancer Res 57, 2384-2387 (1997).
10 Martin, T. A., Watkins, G. & Jiang, W. G. KiSS-1 expression in human breast cancer. Clin Exp Metastasis 22, 503-511, doi:10.1007/s10585-005-4180-0 (2005).
11 McNally, L. R. et al. KISS1 over-expression suppresses metastasis of pancreatic adenocarcinoma in a xenograft mouse model. Clin Exp Metastasis 27, 591-600, doi:10.1007/s10585-010-9349-5 (2010).
12 Nagai, K. et al. Prognostic value of metastin expression in human pancreatic cancer. J Exp Clin Cancer Res 28, 9, doi:10.1186/1756-9966-28-9 (2009).
13 Wang, C. H., Qiao, C., Wang, R. C. & Zhou, W. P. KiSS‑1‑mediated suppression of the invasive ability of human pancreatic carcinoma cells is not dependent on the level of KiSS‑1 receptor GPR54. Mol Med Rep 13, 123-129, doi:10.3892/mmr.2015.4535 (2016).
14 Jayasena, C. N. et al. Plasma kisspeptin: a potential biomarker of tumor metastasis in patients with ovarian carcinoma. Clin Chem 58, 1061-1063, doi:10.1373/clinchem.2011.177667 (2012).
15 Kang, H. S. et al. GPR54 is a target for suppression of metastasis in endometrial cancer. Mol Cancer Ther 10, 580-590, doi:10.1158/1535-7163.MCT-10-0763 (2011).
16 Makri, A. et al. KISS1/KISS1R expression in eutopic and ectopic endometrium of women suffering from endometriosis. In Vivo 26, 119-127 (2012).
17 Ergen, A. et al. Plasma Kisspeptin-54 levels in gastric cancer patients. Int J Surg 10, 551-554, doi:10.1016/j.ijsu.2012.08.014 (2012).
18 Kostakis, I. D. et al. KISS1 and KISS1R expression in gastric cancer. J BUON 23, 79-84 (2018).
19 Li, N., Wang, H. X., Zhang, J., Ye, Y. P. & He, G. Y. KISS-1 inhibits the proliferation and invasion of gastric carcinoma cells. World J Gastroenterol 18, 1827-1833, doi:10.3748/wjg.v18.i15.1827 (2012).
20 Cho, S. G. et al. Kisspeptin-10, a KISS1-derived decapeptide, inhibits tumor angiogenesis by suppressing Sp1-mediated VEGF expression and FAK/Rho GTPase activation. Cancer Res 69, 7062-7070, doi:10.1158/0008-5472.CAN-09-0476 (2009).
21 Curtis, A. E. et al. Kisspeptin is released from human prostate cancer cell lines but plasma kisspeptin is not elevated in patients with prostate cancer. Oncol Rep 23, 1729-1734, doi:10.3892/or_00000818 (2010).
22 Wang, H. et al. Clinical and biological significance of KISS1 expression in prostate cancer. Am J Pathol 180, 1170-1178, doi:10.1016/j.ajpath.2011.11.020 (2012).
23 Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat Methods 19, 679-682, doi:10.1038/s41592-022-01488-1 (2022).
24 Wiederstein, M. & Sippl, M. J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35, W407-410, doi:10.1093/nar/gkm290 (2007).
25 Lamiable, A. et al. PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res 44, W449-454, doi:10.1093/nar/gkw329 (2016).
26 Schrödinger, L., & DeLano, W., . PyMOL, <www.pymol.org/pymol> (2020).
27 Bloodworth, N., Barbaro, N. R., Moretti, R., Harrison, D. G. & Meiler, J. Rosetta FlexPepDock to predict peptide-MHC binding: An approach for non-canonical amino acids. PLoS One 17, e0275759, doi:10.1371/journal.pone.0275759 (2022).
28 PRODIGY: a web server for predicting the binding affinity of protein-protein complexes, <https://wenmr.science.uu.nl/prodigy/> (2020).
29 Systemes, D. BIOVIA Discovery Studio v21.1.0.20298, , <https://www.3ds.com/es/productos-y-servicios/biovia/> (2020).
30 Merrifield, R. B. Solid Phase Peptide Synthesis .1. Synthesis of a Tetrapeptide. Journal of the American Chemical Society 85, 2149-&, doi:https://doi.org/10.1021/ja00897a025 (1963).
31 Taniguchi-Ponciano, K. et al. The KISS1 gene overexpression as a potential molecular marker for cervical cancer cells. Cancer Biomark 22, 709-719, doi:10.3233/CBM-181215 (2018).
Received: 25 June 2023/ Accepted: 26 August 2023 / Published:15 September 2023
Citation: Rodríguez Sarmiento D Y, Toloza Sandoval D J, Rondón-Villarreal P. Structural analysis and cytotoxic evaluation of kisspeptin10 and analogs in types of cancer. Revis Bionatura 2023;8 (3) 61. http://dx.doi.org/10.21931/RB/2023.08.03.61
