2023.08.02.66
Files > Volume 8 > Vol 8 No 2 2023
Molecular detection of Escherichia coli efflux pumps genes isolated from UTI in pregnant women

Wedad Adil Kadhim 1,*, Kareem Ibrahim Mubarak 2,*
1 Diyala University/College of Science/Department of Biology
2 Diyala University/College of Science/Department of Biology
* Correspondence: [email protected], [email protected]
Available from: http://dx.doi.org/10.21931/RB/2023.08.02.66
ABSTRACT
Sixty-three clinical samples from midstream urine samples were collected from pregnant women with urinary tract infections. After microscopic examination, culture and biochemical tests and the final diagnosis using the VITEK-2 system, 25 Escherichia coli isolates were discovered. Antimicrobial susceptibility results showed that E.coli isolates were resistant to gentamicin (%92), amikacin (%28), norfloxacin (%52), Ciprofloxacin (%56), ofloxacin (%60), trimethoprim (%8), chloramphenicol (%80), colistin sulfate (%20), tetracyclin (%68), azithromycin (%48), cefoxitin (%40), amoxicillin-clavulanate (%96), ampicillin (%92). The prevalence of capsule posses, hemolysin production, biofilm formation, and efflux pumps wrer%24,%16,%72 and %44 respectively. The result of efflux pumps genes acrA and acrB gene detection was 100. The acrA and acrB gene expression increased after treatment with the antibiotic Ciprofloxacin. Because of the primary role of Escherichia coli in urinary tract infection and the presence of a high ratio of Multidrug Resistance ( MDR ), and the importance of The efflux pumps in antibiotics resistance, the current study was conducted to determine the MDR isolates from UTI in pregnant women's in Baquba city, the percent of acrA and acrB genes among strains and the effect of Ciprofloxacin treatment on gene expression.
Keywords. Escherichia coli, efflux pumps, acrA gene, acrB gene
INTRODUCTION
Escherichia coli is a gram-negative bacteria, rod-shaped, motile or non-motile, facultatively anaerobic, lactose fermentative1. It is one of the most common types of the Enterobacteriaceae family that naturally inhabit the human gut, making it able to infect other body systems opportunistically when the opportunity arises and cause many diseases 2. E. coli is the leading cause of 90% of urinary tract infections, especially in women, and it comes in second place after respiratory diseases 3. The excessive use of antimicrobial agents led to the problematic treatment of UTI infection due to bacteria having the character of multidrug resistance (MDR) as they can produce many enzymes, such as ESBL Extended Spectrum B-lactamase Enzymes, Metallo B-lactamase MBL and hemolysin 4 bacteria posses other mechanisms that enable them to resist antibiotic, such as changing the target site, changing the permeability of the cellular membrane, inhibiting protein synthesis and efflux pumps. It confers the bacteria multi-resistance to antibiotic 5.
Efflux pumps are transporters of protein located in the cell membrane of bacteria and play a significant role in transporting various substances and expelling them outside the cell to eliminate their harmful effects. It is an essential way for bacteria to develop resistance to antibiotics6. These pumps fall into two main categories based on their genetic origin, which are chromosomal efflux pumps, that is, they carry the genes that code for them on a cell's chromosome, and plasmid efflux pumps carry the genes they encode on portable genetic materials, such as plasmids or transposons 7. There are five types of families of efflux pumps; Major Facilitator Super Family (MFS), Small Multidrug Resistance Family (SMR), Family (Multidrug and Toxic Efflux Family (MATE), ATP - Binding Cassette Family (ABC, Resistance - Nodulation - Division Family (RND)8. The RND family is classified into three classes according to its components, which include: Single-component efflux pumps represented by the protein AcrB located in the inner membrane of the cell, which is encoded by a gene called acrB, which transports hydrophilic antigens. Two-component pumps represented by AcrB in the inner membrane and AcrA lipoproteins in the periplasmic space, encoded by a gene called Acra, Three-component tripartite pumps represented by AcrB in the inner membrane, AcrA proteins in the plasma vacuole, and a funnel-like protein channel called ToIC encoded by the tolC gene), which is located in the outer membrane of a bacterium 9.
Gene expression is known to be an essential feature for maintaining the functional integrity of cells. It occurs in different ways and an organized manner and is divided into positive and negative organizations. Regulation occurs when mRNA begins to be transcribed in prokaryotic mostly. While it becomes more complex when it is present in eukaryotes, so there is more than one mechanism to complete the process of regulation; this process occurs through the association of proteins with a specific sequence on the DNA strand, resulting in either an increase or decrease in the transcription rate in eukaryotic and prokaryotic. In bacteria, this process occurs through the initiation of cloning. An example is the positive and negative control of the lactose operon system operator in E. coli 10.
MATERIALS AND METHODS
Collection of Specimens
Sixty-three urine specimens were collected from pregnant women have symptoms of urinary tract infection admitted to Al-Batool Maternity and Children Teaching Hospital in Baquba City during the period from 1/9/2021- to 1/12/2021.by using disposable collection sterilized container to isolate and identify Escherichia coli from urine specimens and report the information about each patient.
Isolation and identification of bacteria
The urine specimens were inoculated on the MacConkey agar medium after 24 hours plates were inoculated aerobically overnight at 37C֯. According to morphological ( shape and grams stain )and cultural colony characters, the expected isolates were subculture on the Eosine Methylene Blue (EMB) agar, the confirmation of bacterium identification done by biochemical tests including (oxidase, catalase IMVIC tests, urease, Triple sugar iron). Finally, identification is complete by depending on the VITEK-2 compact system 11.
Antibiotic sensitivity test and MIC Determination
The susceptibility E.coli isolates to antibiotics were performed by using 13 antibiotics (gentamicin, amikacin, norfloxacin, Ciprofloxacin, ofloxacin, trimethoprim, chloramphenicol, colistin sulfate, tetracyclin, azithromycin, cefoxitin, amoxicillin-clavulanate, ampicillin) the base of the Disk Diffusion method by spreading bacteria on the Muller-Hinton Agar medium (Vandepitte et al.;2003) the interpretation of results and classify isolates as sensitive and resistance by ( CLSI 2019 )—determination of MIC of ( Ciprofloxacin & Norfloxacin) 12.
Detection of Virulence Factors of E.coli
Several virulence factors of Escherichia coli were detected in bacterial strains, including
Hemolysin production
Bacterial isolates were cultured on plates containing blood agar medium, and the plates were incubated at 37 °C for 22 hours; then, the results were read by observing the type of hemolysis zones around the bacterial colonies13.
Detection of Capsule
The Indian ink method was used for dyeing; take a colony of bacteria and mixed with a small drop of physiological saline using a wooden stick on a clean glass slide; add a drop of Indian ink to the slide and mix, then the slide cover was placed over it so that the liquid spread in a light layer under the glass cover, and examined under the oil lens14.
Detection of Biofilm formation
Phenotypic detection of Biofilm formation, Micro Titer Plate assay according to 15 to detect the biofilm formation, bacteria were inoculated on Nutrient broth medium and incubated at 37°C for 24 hours. After that, the broth cultures were compared with a McFarland Standard using the same medium as were transfer 200 μl of an isolate suspension into each of three wells of a 96-well flat-bottomed polystyrene plate and incubated at37°C for 24 hours. Each well was washed thrice with distilled water (DW) with rough shaking and dried thoroughly. Absolute methanol(200μl) was used for fixation of adhering bacterium. After that, each well was stained with 200μl of 0.5% Crystal Violet for 15 minutes. .washing several times was.to remove the excess stain. Later, the crystal violet associated with adherent cells was retained with 200μl of ethanol per well. The absorbance of wells filled with bacteria-free Nutrient broth served as a negative control. The amount of crystal violet removed by 95% ethanol in each well was quantified by measuring the OD 630 using an ELISA reader. Because. of this, the absorbance values. Represented the intensity of the biofilm formed by well-studied isolates on the surface of the microtiter. The results obtained were categorized into three groups (i.e., non-biofilm producer, moderately and firmly).
Where the absorption of the cultivated pit was compared with the control pits as follows: If OD≤ ODc (Considered non-biofilm producer); if ODc ≤ OD ≤2*ODc (Considered moderately biofilm producer); if 2*ODc ≤ OD( Considered strongly biofilm producer). Where OD (Represent the tested isolates); ODc (Represent control pits).
Phenotypic Efflux pumps detection
Phenotypic Detection of Efflux pumps was conducted by(cartwheel pattern) agar-EtBr method(Martins et al.;2013); dilutions of bacterial were prepared from isolates that revealed the resistance to antibiotics and the ability to biofilm formation after incubated for 24 hours at 24C֯ in the 5 ml of buffer. Their concentration was adjusted by comparing to 0.5 of MacFarland standard solution, then isolates inoculated on tryptic soy agar (TSA) plates containing(0,5,10,15,20,25 µg\ml ) of Ethidium Bromide(EB), the plate was divided to 16 by radial lines(cartwheel pattern), the plates incubated for 16 hours at 37 C֯ in the dark place, the isolates were examined under ultraviolet (UV) trans illuminator to record the fluorescent.
Genotypic detection of efflux pumps
DNA extraction: DNA bacteria was extracted from the isolates of E.coli by using a genomic DNA purification Kit supplemented by the manufactured company (Geneaid, Thailand)16.
Polymerase Chain Reaction (PCR) detected acrAB efflux pump genes (acrA,acrB)in E.coli isolates. The proliferation of acrAB genes with two primer pairs(Bioneer, Korea) Table 1. The amplification of DNA was performed at a volume of 25µl; the reaction mixture contained 5µlAccu powerPCRPremix (Bioneer, Korea), 3µl template DNA, 1.5µl F-primer,1.5µl R-primer and 14µl deionized nuclease-free water. PCR reaction of acrAB genes was performed according to 8 in three steps in Table 2.

Table 1. Sequences and primers concentration used in the study
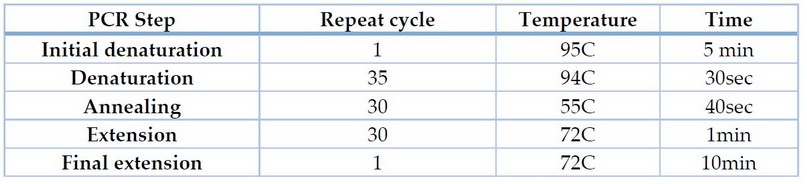
Table 2. Phases of the polymerase chain reaction in the thermal cycler
Gel electrophoresis: after the amplification process in the PCR apparatus, the product was run on 1.5 % agarose gel with 5ml of ethidium bromide in 1x (TBE) buffer using a DNA ladder (100-2000)bp (Bioneer, Korea) at 100 volts for 80 minutes. The visualization of PCR products under 320 nm UV light using a UV transilluminator 12.
Ciprofloxacin Effect on gene expression
RNA Isolation
E.coli Bacteria was pelleted and then lysed in 1 ml of AccuZolTM lysate through the. Two hundred µl of chloroform was added per 1ml of AccuZolTM and shaken vigorously for 15 seconds. The mixture was incubated on ice for 5 minutes. The mixture was centrifuged at 12000 rpm for 15 minutes at 4C, The aqueous phase was transferred to a new 1.5ml tube, and an equal volume of isopropyl alcohol was added. The tubes were mixed by inverting 4 – 5 times and incubated at -20C for 10 minutes. Then Centrifuge at 12000 rpm for 10 minutes at 4C was done, and the supernatant was carefully removed. One ml of 80% ethanol was added and mixed well by vortexing and Centrifuge at 12000 rpm for 5 minutes at 4C was done, then the supernatant was carefully removed, and The pellet was dried; the RNA was dissolved in RNase – free water by passing the solution a few times through the tip of the pipette and stored at -80% until us 18.
Transformation of RNA to DNA
The RNA extracted from each bacterial isolate, primer and water were mixed and incubated at 65°C for 5 minutes, on ice for two minutes. RNA was 10pg-500mg, random primer 1µl, 2XES reaction mix 10µl, RT\RL enzyme 1µl, gDNA remover 1µl, RNase- free water 20µl (Bioneer, Korea). The mixture was treated with 25c֯ for 10 minutes,42c֯ for 15 minutes, and 85c֯ for 5 seconds to inactive enzymes.
Real-time quantitive reverse transcription (qRT-PCR)
RT-qPCR is performed in a reaction solution of 20µL volume using BrightGreen qPCR MasterMix by mixing the 0.6 µL of primer, 10µL of BrightGreen 2X qPCR master mix, ≤ 500 ng of cDNA, Nuclease-free H2O to 20 µl.
The generated solution was placed in Real-time PCR Cycler for thermal reaction to measure the Cycle Threshold (CT) value. RT-PCR is used for quantification of the levels of gene expression. The measured CT values during the thermal reaction are recorded to the computer in the following measurements 19.
Real-Time qRT-PCR analysis: ΔCT (test) = CT gene of interest (target, test) – CT internal control, ΔΔCT=ΔCT (test)- ΔCT (calibrator), 2-ΔΔCt = Normalized expression ratio(20).
Statistical analysis
Statistical analysis was carried out using the SPSS version 20, and the results were analyzed using the Chi-Square test with a probability level of P≥ 0.05 21.
RESULTS
The diagnosis results showed that the number of E.coli isolates obtained from a total of 63 urine specimens was 25. Biochemical tests reveal that all the isolates of E.coli were positive for catalase, Indole, and Methyl Red and negative for Oxidase, Vogues-Proskauer, citrate utilization and Urease, TSI(A\A, H2S+, gas+).E.coli isolates were gave pink colonies on MacConkey agar, dark blue with green metallic sheen on(EMB)agar.
Sensitivity of E.coli to Antibiotic
The results of the current study showed the resistance ratio to Gentamicin, Tetracyclin, and Trimethoprim were 23(%92),17(%68), and 2(%8), respectively.
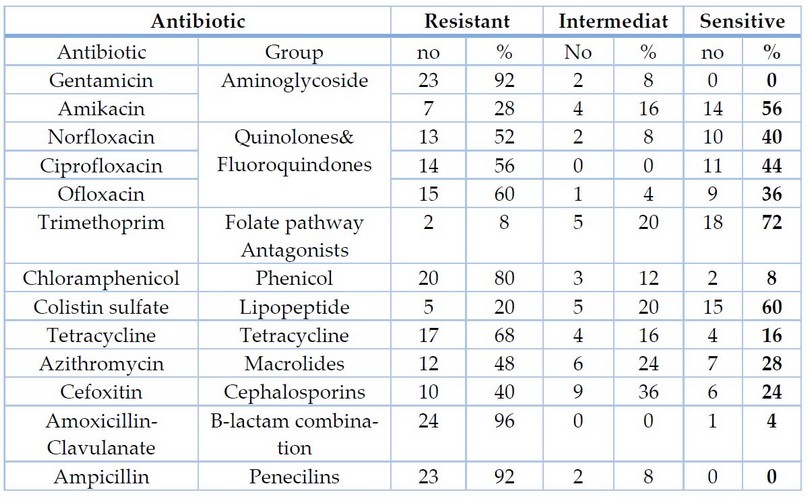
Table 3. Sensitivity of Escherichia coli isolates to antibiotics.
MIC Determination
The current study revealed a contrast in the MIC values of E.coli isolates against the antibiotics ciprofloxacin and norfloxacin for isolates of pregnant women infected with urinary tract infections. Where it ranged between (8-1024) µg/ml for Ciprofloxacin and (8-512) µg/ml for norfloxacin, and the range of MIC for the four isolates (2, 5, 30, 24) were treated to study the gene expression reach(8-256)µg\ml.
Hemolysin production: The results of the current study showed that 4(%16) of E.coli produced hemolysin.
Capsule posses: The results of the current study showed that 6 (%24) of E.coli possess a capsule. This result approached 26, who found that %37.2 of E.coli isolates are capsule posses.
Biofilm formation: efflux pumps production: The results of the current study showed that 11(%44) of E.coli produced efflux pumps.
Detection of (acrAB)genes: molecular detection indicates that all isolates carrying acrAB genes by %100 were carriers Figure 1,2.
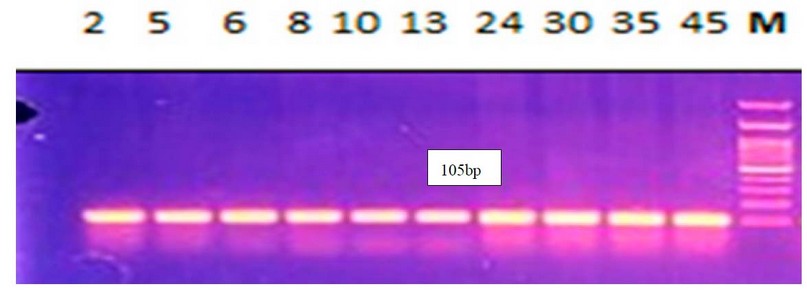
Figure 1. Electrophoresis of amplified acrA(105bp) for E.coli by 1.5% agarose gel, using DNA Ladder 100-2000bp, 80V\cm for 80 min, stained with ethidium bromide dye and visualized under UV transilluminator documentation All lanes are shown to have the gene.
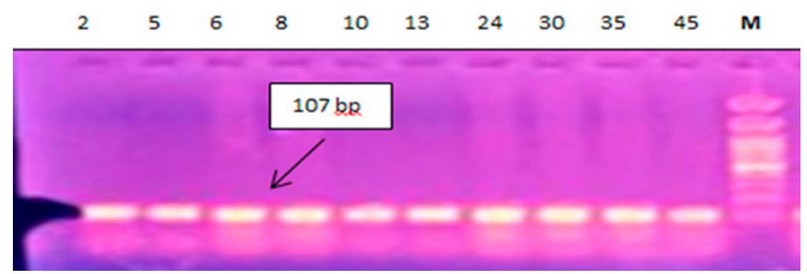
Figure 2. Electrophoresis of amplified acrB (107bp) for E.coli by 1.5% agarose gel, using DNA Ladder 100-2000bp, 80V\cm for 80 min, stained with ethidium bromide dye and visualized under UV transilluminator documentation All lanes are shown to have the gene.
Ciprofloxacin Effect on Gene Expression
The results of the current study for four bacterial E.coli of isolates (2, 5, 30, 24) isolated from pregnant women before and after treatment with Ciprofloxacin using the qRT-PCR technique. Using the primers described in Table 1. showed amplification of the acrA gene with a value of threshold cycle(CT ) before isolates (2,5,30,24) treated with sub-MIC CIP, Where recorded (31.62, 30.42, 27.79, 28.99). The quantitative changes in mRNA were determined using an equation Threshold limit Ct (2-△△ct), as shown in Table 4, after treatment of the isolates with CIP. Isolates (2, 5) showed high genetic expression after treatment with cip where recorded (3.9737, 9.0630). As for the (30, 24), It was not genetically expressed after treatment with cip Table 4.
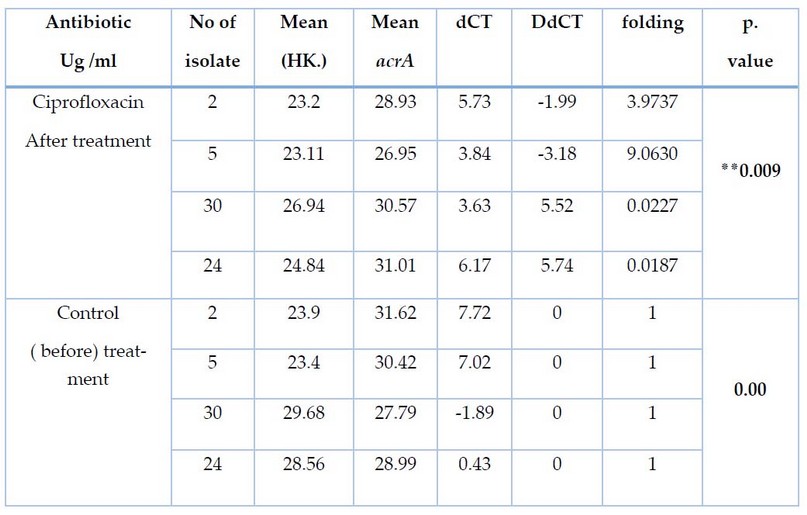
Table 4. Effect of ciprofloxacin treatment on CT and Fold values of acrA gene expression for E.coli
Results of the current study for four bacterial E.coli of isolates (2, 5, 30, 24) isolated from pregnant women before and after treatment with Ciprofloxacin using qRT-PCR technique. Using the primers described in Table 1. showed amplification of the acrB gene with a value of threshold cycle(CT ) before isolates (2,5,30,24) treated with sub-MIC CIP, Where recorded (22.75, 22.65, 24.7, 28.3) the quantitative changes in mRNA were determined using an equation Threshold limit Ct (2-△△ct), as shown in Table 5. After treatment of the isolates with CIP. Isolates (2, 5) showed high genetic expression after treatment with cip where recorded (5.3889,4.3771). As for the (30, 24), It was not genetically expressed after treatment with cip Table 5. Our study agreed with (Ma et al;1995), which showed increased gene expression for acrAB after treatment with cip.
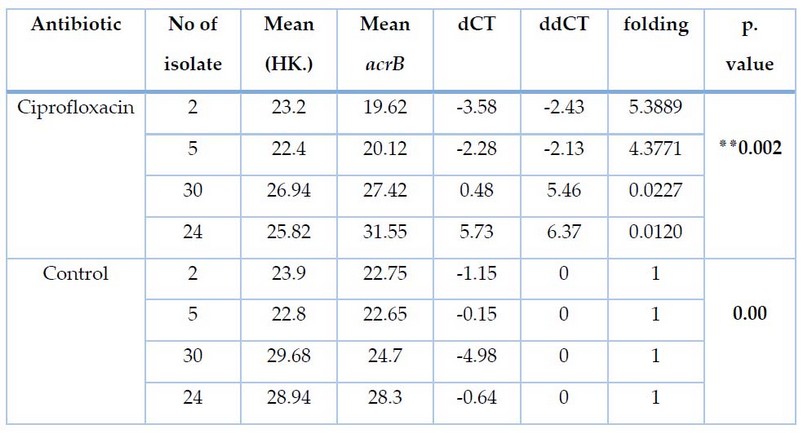
Table 5. Effect of ciprofloxacin treatment on CT and Fold values of acrB gene expression for E.coli
DISCUSSION
The results of the current study showed the resistance ratio to Gentamicin, Tetracyclin; Trimethoprim was 23(%92),17(%68), and 2(%8), respectively. This result did not agree with Jazayeri 22, which showed that the resistance to the antibiotics were%70,%30,%40 respectively, while the resistance ratio of E.coli isolates to Amoxicillin-Clavulanate was 24(%96). This result was similar to 22, which showed that the resistance to the antibiotics were%96, while the resistance ratio of E.coli isolates to Ciprofloxacin was 14(%56). This result was similar to 23. the resistance ratio of E.coli isolates to Ampicillin was 23(%92). This result did not agree with 23, which showed that the resistance to the antibiotic % was 81.7. the resistance ratio of E.coli isolates to chloramphenicol and norfloxacin were20 (%80). this result did not agree with 24. which showed that the resistance to the antibiotics were%42.whil the resistance ratio E.coli isolates to Amikacin was 7(%26).this result was approached to 24. which showed that the resistance to the antibiotic was %21.whil the resistance ratio E.coli isolates to Cefoxitin10 (%40). This result was approached to 25, showing that the antibiotic resistance was 42%. The resistance ratio E.coli isolates to ofloxacin15 (%60). Another study does not agree with the current study, where resistance to the antibiotic was %39.6 25. The current study showed that 4(%16) of E.coli produced hemolysin. This result does not agree with 24, who found that %41.9 of E.coli isolates are hemolysin-producing. The current study showed that 18 (%72) of E.coli formated biofilm. This result approach 27, who found that %90 of E.coli isolates are biofilm formation. The results of the current study showed that 11(%44) of E.coli produced efflux pumps. This result does not agree with 27, who found that %70 of E.coli isolates are efflux pump production. Moreover, molecular detection indicated that all isolates carrying acrAB genes by %100 were carriers; the results agreed with the results of 27.
CONCLUSIONS
The results of isolation and diagnosis of Escherichia coli bacteria showed multi-resistance to antibiotics due to their possession of efflux pumps. Moreover, after treatment with ciprofloxacin antibiotic, the results of the gene expression of acrA and acrB genes recorded high expression in pregnant women.
REFERENCES
1. Jaweetz E, Melnick J A and Adelberg E A. Review of Medical Microbiology 27th ed. McGraw-Hill Higher Education. 2016; pp 850.
2. Nascimento, J. A., Santos, F. F., Valiatti, T. B., Santos-Neto, J. F., M Santos,A. C., Cayô, R., ... & AT Gomes, T. Frequency and Diversity of Hybrid Escherichia coli Strains Isolated from Urinary TractInfections. Microorganisms, 2021; 9(4), 693. 13.
3. ALobaidy, B. .; Al-Falahi, M. N. .; Almarie, A. A. . Effect Of 6-Bap Growth Regulator On Seed Priming Of Several Bread Wheat Varieties Under Water Irrigation Salinity Stress. JLSAR 2021, 2, 21-26..
4. Ye, Q., Wu, Q., Zhang, S., Zhang, J., Yang, G., Wang, H., ... & Wang, J. antibiotic-resistant extended spectrum ss-lactamase-and plasmid-mediated AmpC-producing Enterobacteriaceae isolated from retail food products and the pearl river in Guangzhou, China. Frontiers in microbiology, 2017; 8, 96. 17.
5. Kapoor, J.; Saigal, S. and Elongavan, A. Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. J Anaesthesiol Clin Pharmacol. 2017; 33(3):300-305. 8.
6. Zhi Li, X.; Elkins, C. A. and Zgurskaya, H. I. Efflux-Mediated Antimicrobial Resistance in Bacteria Mechanisms, Regulation And Clinical Implications. International Publishing. Switzerland. 2016; 848 pp.
7. Puzari, M. and Chetia, P. RND efflux pump mediated antibiotic resistance in GramNegative Bacteria Escherichia coli and Pseudomonas aeruginosa: A Major Issue Worldwide. World J Microbiol Biotechnol. 2017; 33(24): 1-8. 14.
8. Maleki, D.; Jahromy, S. H.; Karizi, S. Z. and Eslami, P. The prevalence of acrA and acrB Genes Among Multiple-Drug Resistant Uropathogenic Escherichia coli Isolated from Patients with UTI in Milad Hospital, Tehran. Avicenna J Clin Microb Infec. 2017; 4(1): 1-7. M
9. Hayashi, K.; Nakashima, R.; Sakurai, K.; Kitagawa, K.; Yamasaki, S.; Nishino, K. and Yamaguchi, A. AcrB-AcrA Fusion Proteins That Act as Multidrug Efflux Transporters. J Bacteriol. 2016; 198(2):332–342. 7.
10. Maysaloon W. Ibraheem, Abdulkhaliq A. Farhan, Sataa M. Salih, Th.T. Mohammed. Carcass characteristics of Awwasi lambs supplemented with Selenium and Vitamin D3. Iranian Journal of Ichthyology, 2022; 9(1) : 355-359.
11. Wanger A, Chavez V, Huang R S P, Wahed A, Actor J K and Dasgupta A. Microbiology And Molecular Diagnosis In Pathology. Elsevier Inc.All Rights Reserved. 2017; 300pp.
12. Moerllo , J.A. ; Granto , PA and Mizer , H.E. LaboratoryManual and workbook in Microbiology : Applications to patient Care. 2003.
13. Tille, P.M. Baily And Scott's Diagnostic Microbiology. 41thed. Elsevier,Inc. China. 2017; 1115pp.
14. Atlas, R. M. ; Brown, A. E. and Parks, L. C. Laboratory Manual Experimental Microbiology. 1st. ed. Yearbook , Mosby Inc. 1995;148: 884-8.
15. Fusco, A.; Coretti, L.; Savio, V.; Buommino, E.; Lembo, F. and Donnarumma, G. Biofilm Formation and Immunomodulatory Activity of Proteus mirabilis Clinically Isolated Strains. Int J Mol Sci. 2017;18(2): 1-11.
16. Sambrook J and Russell D. Molecular Cloning Laboratory Manual. 3rd ed. Cold Spring Harbor, New York. USA. 2001.
17. Lee,P.Y.,Costumbrado,J.,Hsu,C.Y.,&Kim,Y.H. Agarose gel electrophoresis for the separation of DNA fragments . Journal of visualized experiments:JoVE, 2012 ; (62).1-6.
18. Chomczynski, P., & Sacchi, N. The single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction: twenty-something years on. Nature protocols, 2006; 1(2), 581- 585.
19. Abtan, J., Bhatt, D. L., Elbez, Y., Sorbets, E., Eagle, K., Reid, C. M., ... & REACH Registry Investigators. Geographic variation and risk factors for systemic and limb ischemic events in patients with symptomatic peripheral artery disease: Insights from the REACH Registry. Clinical cardiology, 2017; 40(9), 710-718.
20. Livak, K. J., & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods, 2001; 25(4), 402-408. 10.
21. Levesque R. SPSS Programming and Data Management. 4th ed. Chicago. 522. 2007.
22. , M. A., & IRAJIAN, G. R. Asymptomatic urinary tract infection in pregnant women. 2009.
23. Demilie, T., Beyene, G., Melaku, S., & Tsegaye, W. Urinary bacterial profile and antibiotic susceptibility pattern among pregnant women in North West Ethiopia. Ethiopian journal of health sciences, 2012; 22(2).
24. Mishra, S., Mohapatra, S., Ranjit, M. R., & Panda, G. K. Biofilm Formation as a Uropathogenic Marker of E. Coli Isolates Causing Urinary Tract Infections among Pregnant Women.
25. Thapa, R., Lamichhane, P., Banjara, M. R., & Acharya, G. P. Prevalence Of Extended Spectrum Beta Lactamase Producing Uropathogens In Pregnant Women. Prevalence, 8(1). Jazayeri, M. A., & Irajian, G. R. (2009). Asymptomatic Urinary Tract Infection In Pregnant Women. 2015.
26. Olorunmola, F. O., Kolawole, D. O., & Lamikanra, A. Antibiotic resistance and virulence properties in Escherichia coli strains from cases of urinary tract infections. African journal of infectious diseases, 2013; 7(1), 1-7.
27. Al-Saadi, Z. H. A., & Abdullah, R. M. Phenotypic And Molecular Detection Of Escherichia Coli Efflux Pumps From Uti Patients. Biochemical And Cellular Archives, 19(Suppl. 1), 2371-2376. 2019.
Received: May 15, 2023/ Accepted: June 10, 2023 / Published: June 15, 2023
Citation: Kadhim, W.A.; Mubarak, K.I. Molecular detection of Escherichia coli efflux pumps genes isolated from UTI in pregnant women. Revis Bionatura 2023;8 (2) 66. http://dx.doi.org/10.21931/RB/2023.08.02.66