2023.08.04.61
Files > Volume 8 > Vol 8 no 4 2023
New Records in Iraq and Arab Nations for some Fungi Isolated from Al-Barakia wastewater treatment plant in Al-Najaf Province
Nihad Mutlag1*, Douaa Hussain 2
2University of Kufa / Najaf/ Iraq;
*Correspondence: [email protected]
Available from. http://dx.doi.org/10.21931/RB/2023.08.04.61
ABSTRACT
This study was conducted in 2020 in the wastewater treatment plant in Al-Barakia Najaf, where samples were taken in November from the Bioshft unit and the compact unit from the plant to know the efficiency of the plant in treatment. The process of isolation and purification was Microbiology Laboratory_ Ecology and Pollution Department - College of Science - University of Kufa. Its diagnosis was carried out at the Plant Virus Laboratory - College of Agriculture - the University of Karbala, and (19) fungal isolates isolated from wastewater treatment plants were diagnosed. These isolates were analyzed using the PCR technique and determination of the nucleotide sequences of the polymerase chain reaction products that were amplified from those isolates using ITS4 and ITS1 primers. It belonged to the fungus A. caespitosus. and isolated A flavus (7,8), T. asperellum (9-11) A.tubingensis (12), A.terrus(13), A.niger (14,15), A. alternata (16), C.sphaerospermum (17), A.oryzae (18), Acremonium sp (19), similarity rate of 100% with isolates registered with the NCBI. The results show that all the isolated fungi diagnosed in this study are recorded globally. However, they are not recorded in Iraq, and (5) isolates are not recorded in the Arab nation and Iraq, including A. tubingensis, C.sphaerospermum, A. alternate, and A.oryzae, while the isolate Acremonium sp. According to the National Center for Biotechnology Information NCBI, it is registered only in Germany under the number (AJ557731), and the similarity rate is 100% with the isolate diagnosed in this study.
Keywords: wastewater; Molecular Identification; fungi; Polymerase chain reaction (PCR); DNA sequence analysis.
INTRODUCTION
Water covers about 71% of the Earth's surface and makes up 65% of our body. We all want cleaning water - to relax, drink, and enjoy. If polluted, this water loses economic and aesthetic value, and it can pose a significant hazard to health and aquatic life. Such as the fish that live in it and the wildlife that depends on it1. One of the more serious environmental issues is the chemical pollution of rivers and lakes. Because chemical pollution is carried by water and enters rivers and streams, it causes massive destruction2,3. Although some types of water pollution may occur through natural processes, they are primarily the result of human activity. Humans use water every single day in industry and homes. Used water is taken from lakes, rivers, and underground; after use pollution, most of the water returns to the same sites. This use of water is called "wastewater." Water not treated before is released into rivers, leading to severe pollution 1. Wastewater is a mixture of water-borne effluents disposed of through establishments, dwellings and industrial facilities, as well as groundwater and surface water4. This water generally contains a large amount of waste that requires an amount of oxygen and pathogens (microbes), Organic matter, plant growth-stimulating nutrients, minerals, sediments and inorganic chemicals. The water may also contain toxic compounds 5. Wastewater can be defined as a mixture of water-borne effluents discharged from dwellings, commercial and industrial establishments, and institutions, along with groundwater, rainwater, and surface water. Generally, it contains a large amount of waste that requires oxygen, pathogens or pathogens, organic matter, nutrients for plant growth, chemicals, sediments, inorganic materials, and minerals. It may also include toxic substances. 1
Wastewater is divided into four types:
1. Household sewage: wastewater emitted from business establishments, households, and similar facilities; Industrial sewage: containing a variety of wastes
2. Leakage or/Outflow: Water leaks into the sewage system via indirect or direct means like leakage connections
3. Porous or cracked walls Storm water that enters the sewage system through rainfall drainage connections
4. Rainwater: runoff resulting from floods as a result of rainfall. For several years, the main objective of wastewater treatment in cities has been to lower the concentration of suspended particles, oxygen-requiring chemicals, dissolved substances, and potentially dangerous bacteria and fungi. However, in recent times, there has been increased emphasis on improving the ways of solid waste disposal from municipal treatment operations. Wastewater is the primary storage location for all microorganisms, including fungi and heavy metals. 6,7. Wastewater is the primary reservoir for every microorganism, including fungal ones 8,9. Fungi are eukaryotic microorganisms in a separate kingdom; about 95% of these species are currently unknown and must be discovered—discovery 10,11. The classification of fungi based on their morphological characteristics may often give accurate results. However, many researchers do not rely on morphological characteristics because it may require sufficient experience in the field of classification, especially since there are groups of fungi that are very similar and require significant effort and time. It is also often an inaccurate method. After all, it is affected by environmental factors that may influence the shape and colors of spores, size and fungal colonies 12,13. PCR technology is the molecular technique that relies on the selection and amplification of a specific area of the organism's genome, and thus knowledge of the genetic relationships in terms of similarity and the difference between the species of fungi that will help in the phenotypic diagnosis of the study fungi 14,15. This study aimed to isolate and diagnose fungi isolates using (PCR), determine the sequence of nitrogenous bases, and identify similarities and differences in genetics between the isolates—compared with the same fungi internationally diagnosed recorded in ( NCBI).
MATERIALS AND METHODS
Al-Barakia wastewater treatment plant is located in Najaf Governorate to the southeast of the city of Kufa on the Euphrates River (Shatt al-Kufa) at the coordinates (N 32 00 40, E 44 25 20) and as shown in Figure (1): The wastewater treatment plant consists of two projects, where samples were collected from the modern project, which contains two treatment units, Bio_Sheft unit, and compact unit. Water samples were collected from the Al-Barakah sewage treatment plant in the morning of November 2020, and (3 replicates) were taken for the Biosheft unit and the Compact unit. A sample was taken from the al-Kufa River (1 km) before entering the site to carry out laboratory tests, isolate and diagnose fungi from the sewage station, where they are collected using polyethylene containers with a capacity of (5) liters that are washed with distilled water before use. According to (Himedia), PDA was prepared for the isolate of fungi, as (39 g) PDA powder was dissolved in 1 liter of "distilled water" and autoclaved at 121 °C, then 15 ib.ng-1 pressure. The media is then cooled, and an antibacterial, chloramphenicol (250) mg/liter, is added to stop bacterial growth before pouring.
Furthermore, the fungi are isolated and purified. A separate method (1 ml) is taken from water samples for the two sites and placed in Petri dishes, 15 ml of PDA medium is added, and three replicates are taken for each site. The media is left until the PDA hardens and then incubated at 25 -+ 28 °C for five days until the microbial colonies grow correctly. The process of purification and diagnosis was carried out, including morphology and microscopic features of the fungi.
Molecular diagnosis of fungi isolates
The DNA extraction technique of fungi isolates, by the method described by the American company Zymo0 Research, was used, with the kit (Cat. No. D6005) provided by the same company.
Polymerase chain reaction (PCR)
To diagnose isolates fungus isolated in this study, a test polymerase chain reaction(PCR) through the use of the Kit (Maxime PCR PreMix (i-Taq), Cat. No.25026), made by the Korean company (iNtRoN). They performed a (PCR) with total volumes of the (20μl), which contained (1μl) of each initiator's front (ITS1:TCCGTAGGTGAACCTGCGGG) and posterior (TCCTCCGCTTATTGATATGC: TS4) (White et al.,1990) as well as (1 μl) of DNA extracted. All of the above components were placed inside a tube provided by the manufacturer, and the volume was 20 μl of nuclease-free water. The subsequent steps have doubled the DNA of the fungal isolates. PCR conditions are: DNA denaturation first for five minutes at 98°C, followed by the final denaturation process. that consists of 35 cycles during 40 seconds at 94°C, and the primer annealing process continues during period40s at 55 °C, then the initial elongation by the PCR, amplified products during the period (1 minute )at 72°C. Elongation step at 72°C. Finally, the PCR ended by one step with the final elongation at 72°C 16.
DNA sequence analysis for fungi
This study aims to diagnose the fungal isolates (1-19) and those isolated from the Al-Barakia wastewater treatment plant by PCR. Therefore, the primer (ITS1 and ITS40) was delivered to the Macrogen Company of South Korea to determine The sequence of the nitrogenous bases. All nitrogenous base sequences are analyzed by BLAST to compare the results with the facts provided in NCBI) belonging to the like fungus at (NCBI) Phylogenetic trees analysis is using drawn MEGA X17.
RESULTS
Isolation and Identification of Fungi
Table (1) identifies 19 fungal isolates from the Al-Baraka wastewater treatment plant in Najaf. The fungi were identified by molecular diagnosis by PCR technique, and the most fungi were Aspergillus (13) isolates, Trichoderma (3) isolates, A. alternate (1), Acremonium sp (1), and C. sphaerospermum((1) isolate and the similarity and difference ratios were compared with fungi isolates previously registered in NCBI (BLAST) as shown in Table 1
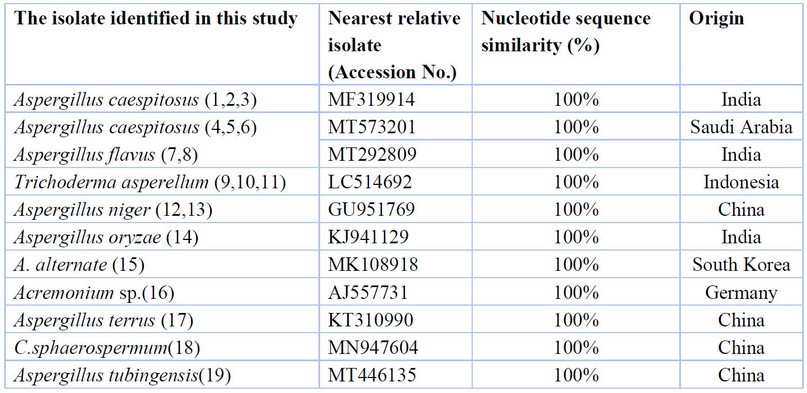
Table 1. Fungi isolated from wastewater treatment plant.
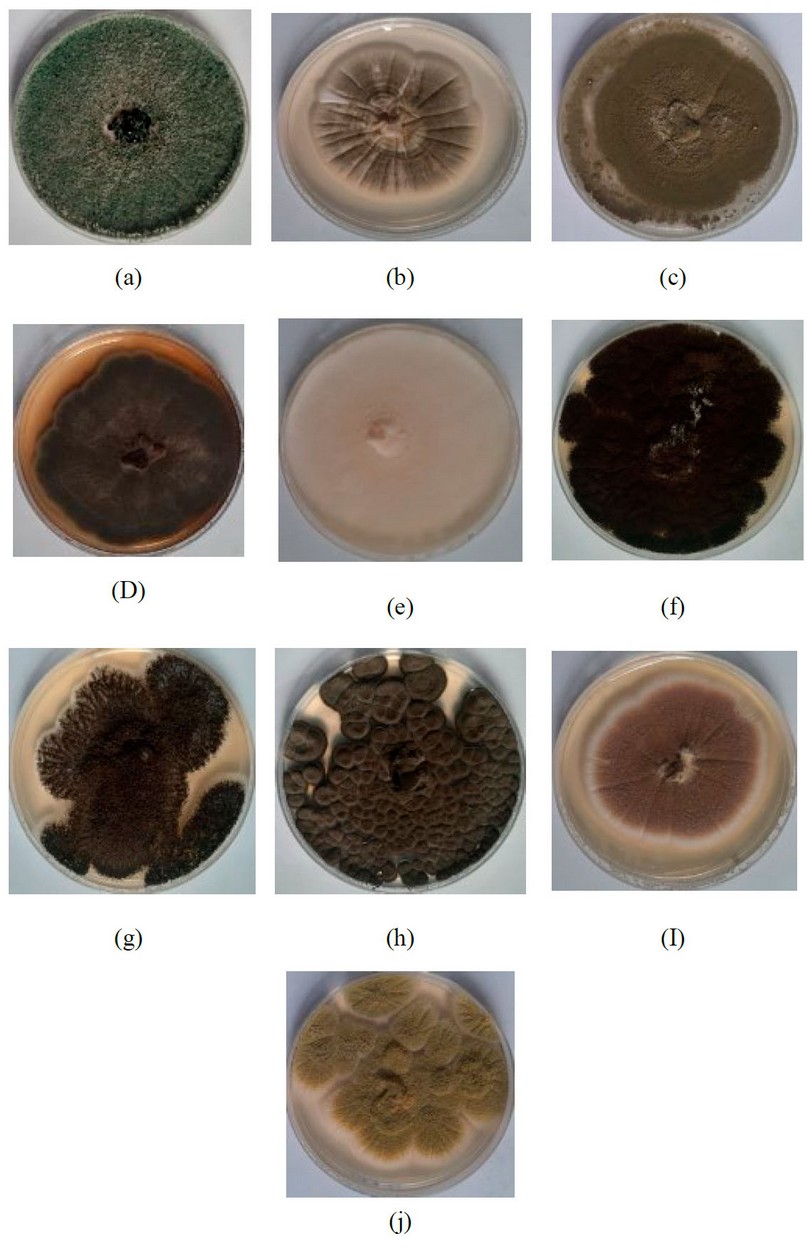
Figure 1. (a) Trichoderma asperellum. (b) Aspergillus caespitosus.(c) Aspergillus flavus (d) Alternaria alternate. (e) Acremonium sp. (f) Aspergillus niger (g) Aspergillus tubingensis (h) Cladosporium sphaerospermum (I) Aspergillus terrus (J) Aspergillus oryzae.
Molecular Diagnostics of Fungi by Using PCR Technology
The results of DNA extraction from fungal isolates (1-19) and PCR exposure revealed (Figure 3.2) the capability of multiplying PCR-amplified product with sizes ranging from (620 bp) and using the pair of primers (ITS1) and (ITS2) (ITS4). M = DNA ladder marker (100bp DNA ladder marker with volume each fastened to the number of nitrogen base pairs (bp) on the Figure's left side). N.C. refers to unfavorable Comparison (a mixture containing PCR components without DNA addition). as shown in Figure 2.
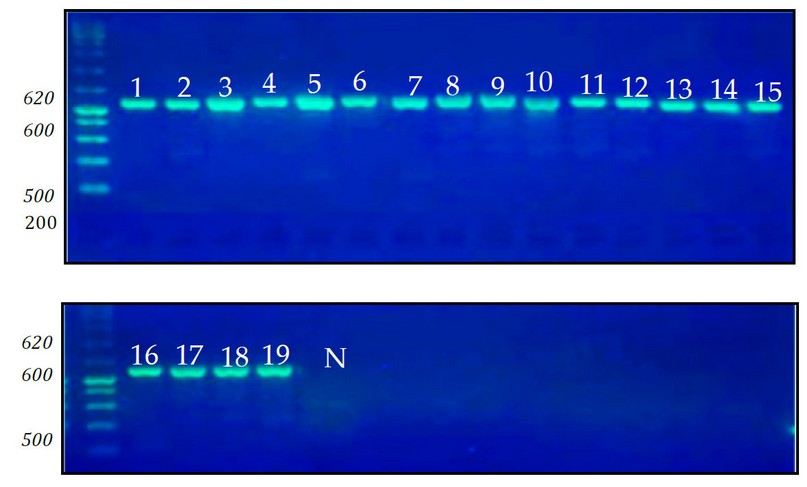
Figure 2. DNA products amplified by polymerase chain reaction (PCR) from fungi isolated (1-19) from the wastewater treatment plant.
The nucleotide sequence analyses of the multiplexed DNA produced by the fungal isolate were shown in the results. Making utilization of (the BLAST) program proves that all the fungal isolates belong to the fungus A caespitosus. Analysis results showed the nucleotide sequences generated from the identified. A. caespitosus isolates. The similarity rate was 100%. Among isolates (1, 2, 3), the similarity percentage was 100% between isolates (4, 5, 6), while the similarity percentage was 99% between isolates (1, 2, 3) and (4,5 and 6) diagnosed in this study. As shown in Figure (3)
A. caespitosus

Figure 3. Nucleotide sequence alignment of the PCR products amplified by (PCR) from A. caespitosus isolates (1, 2, 3, 4, 5, and 6) isolated in this study.
The numbers on the right side of the fungi name refer to the sequences of nitrogen bases in the DNA products of the fungi in this experiment.
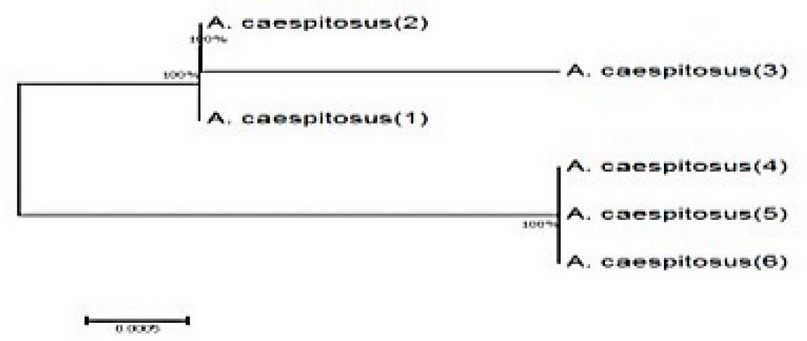
Figure 4. A phylogenetic tree shows the genetics between the A. caespitosus isolates (1, 2, 3, 4, 5, and 6) isolated from the wastewater treatment plant.
Isolation of A.caespitosus * fungus isolates in this study.
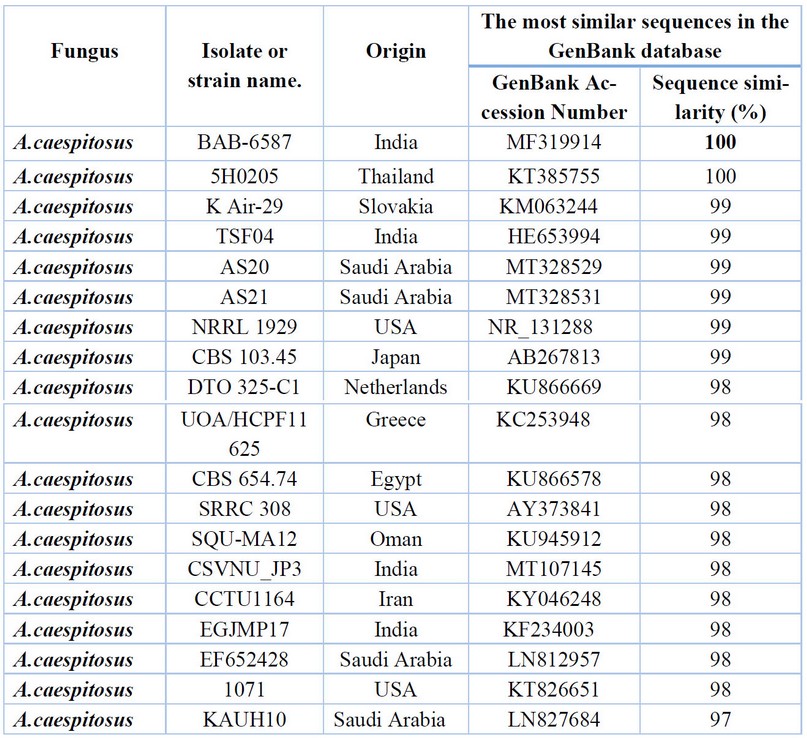
Table 2. Comparison between the similarity ratios of Aspergillus caespitosus isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
Isolation of A.flavus * fungus isolates in this study.
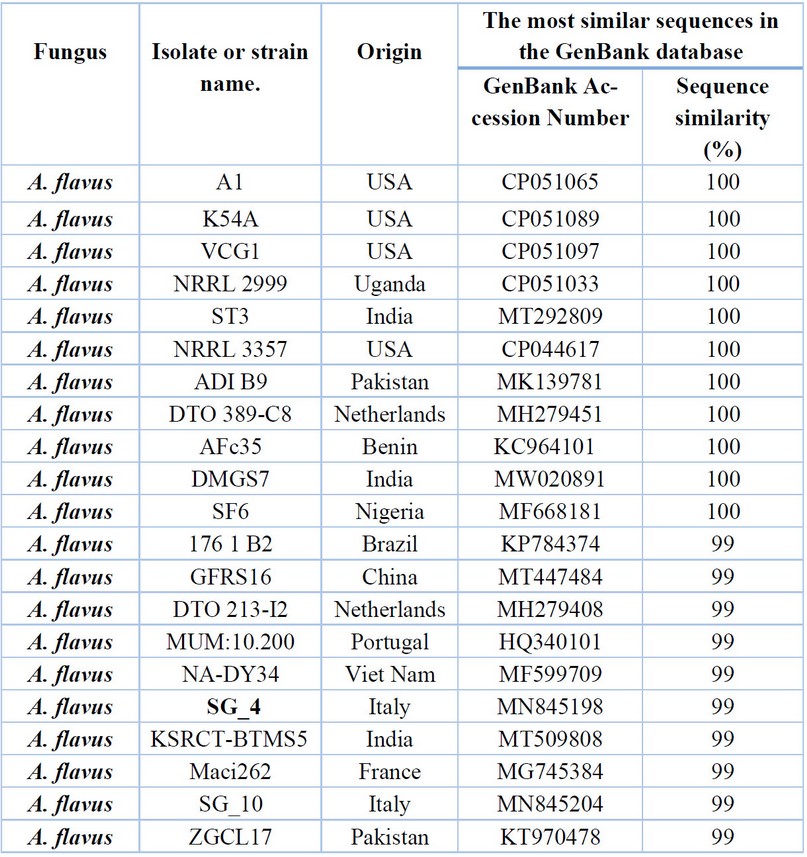
Table 3. Comparison between the similarity ratios of Aspergillus flavus isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
The result showed that using the BLAST program, the nucleotide sequence analyses of the PCR-amplify-produced fungal show that all isolates belong to Trichoderma asperellum—analysis results of the nucleotide sequences generated from the identified. Trichoderma asperellum isolates show. The similarity isolates between (9, 10, and 11) are 100%. They were diagnosed in this study (Figure 5).
Trichoderma asperellum
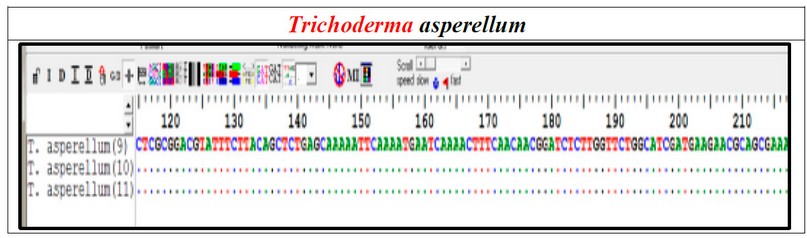
Figure 5. Nucleotide sequence alignment of the PCR products amplified by (PCR) from T. asperellum isolates (9,10 and 11) isolated in this study.
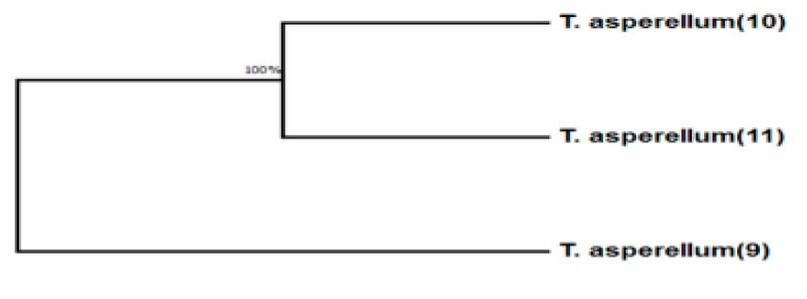
Figure 6. The phylogenetic tree shows the genetic relationships of T.asperellum (9, 10, and 11) isolates. This experiment isolates it from the wastewater treatment plant.
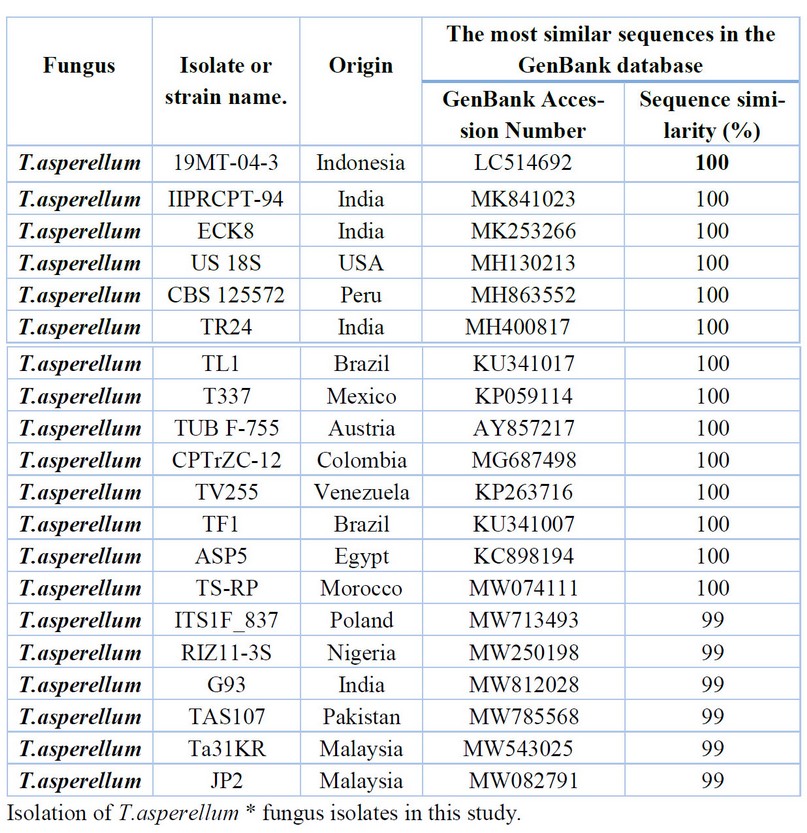
Table 4. Comparison between the similarity ratios of Trichoderma asperellum isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
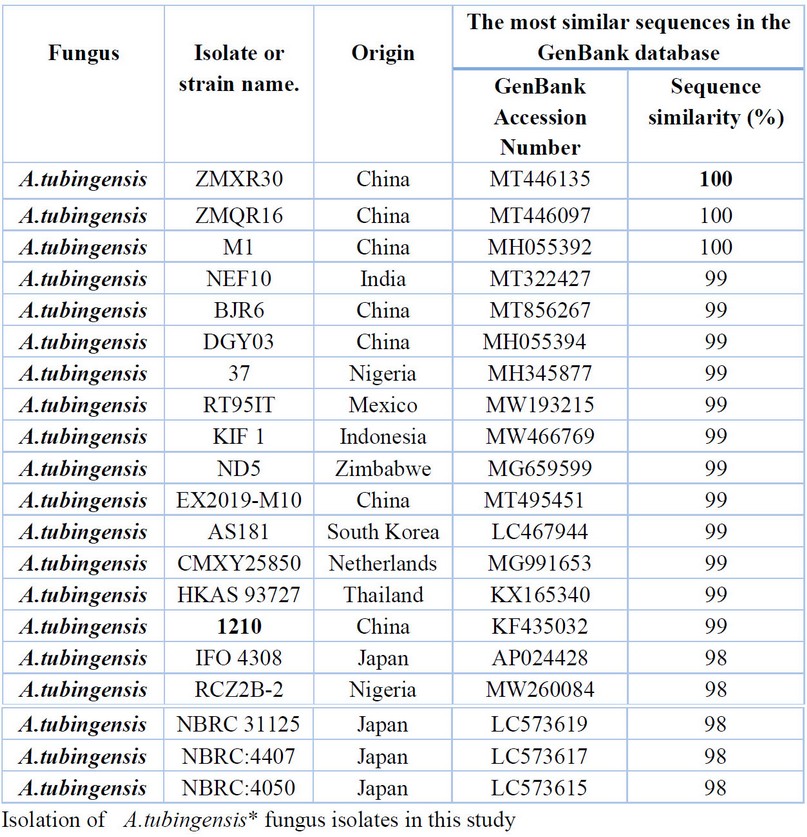
Table 5. Comparison between the similarity ratios of Aspergillus tubingensis isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
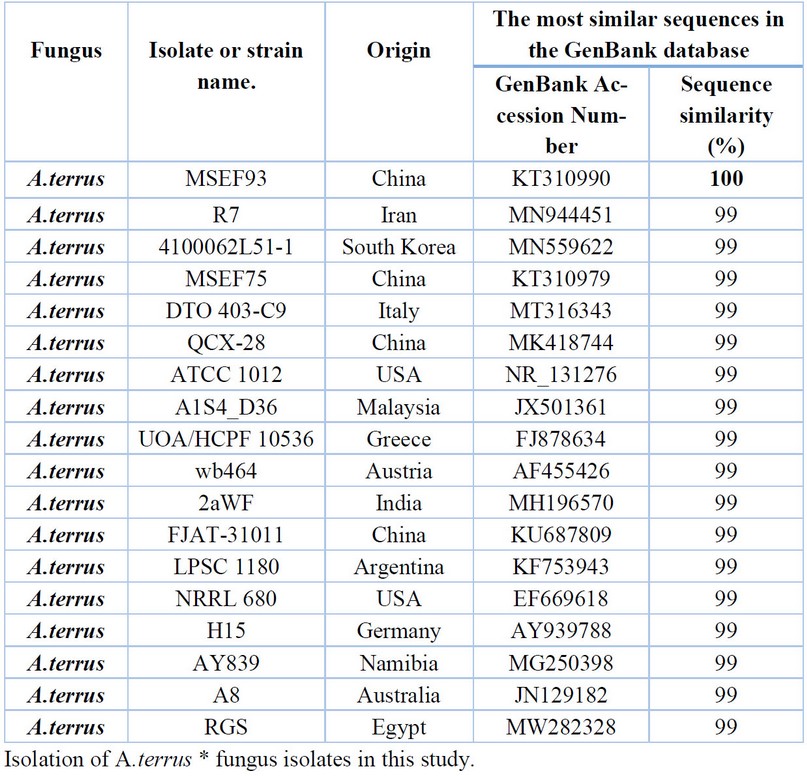
Table 6. Comparison between the similarity ratios of A.terrus isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
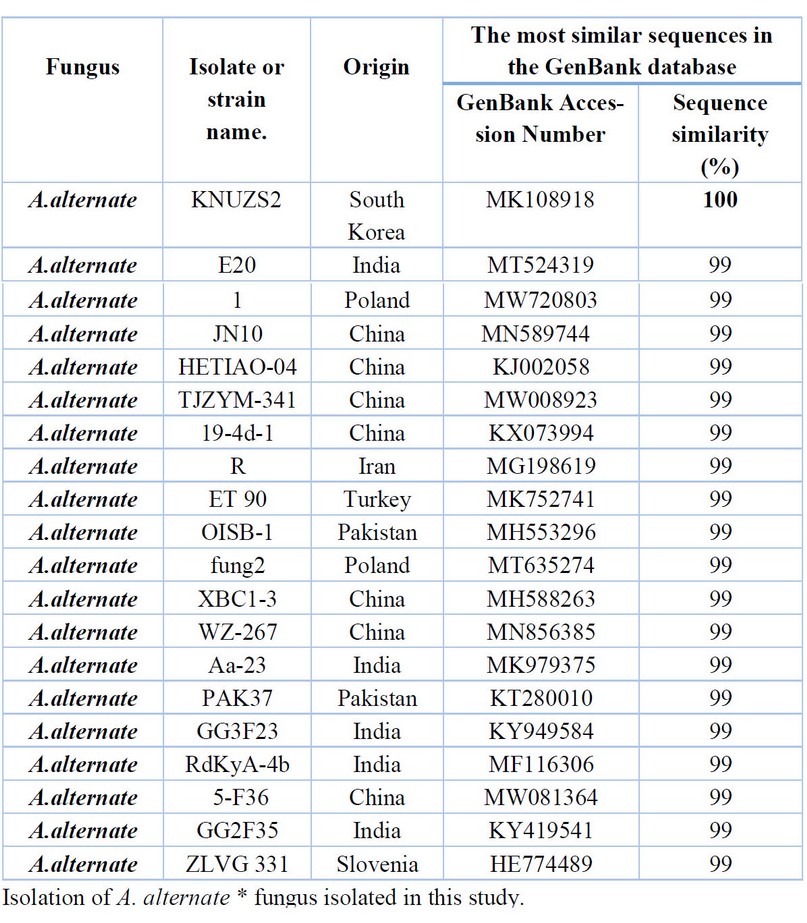
Table 7. Comparison between the similarity ratios of Alternaria alternata isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
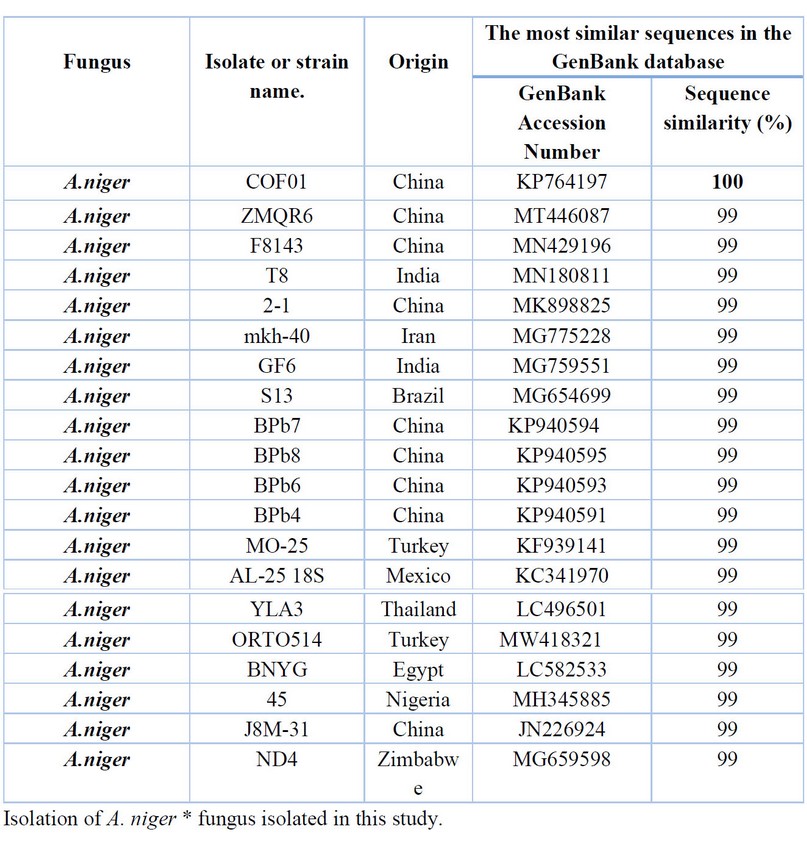
Table 8. Comparison between the similarity ratios of Aspergillus niger isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
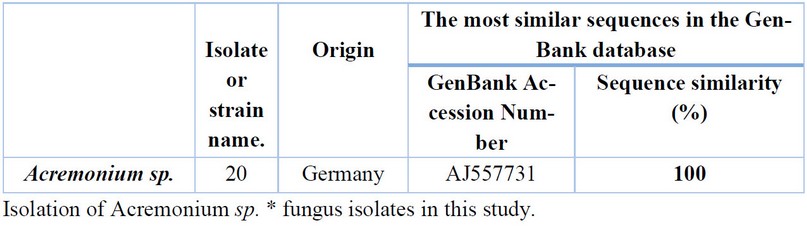
Table 9. Comparison between the similarity ratios of Acremonium sp isolates from a wastewater treatment plant with the other isolate of the same fungus previously recorded in NCBI.
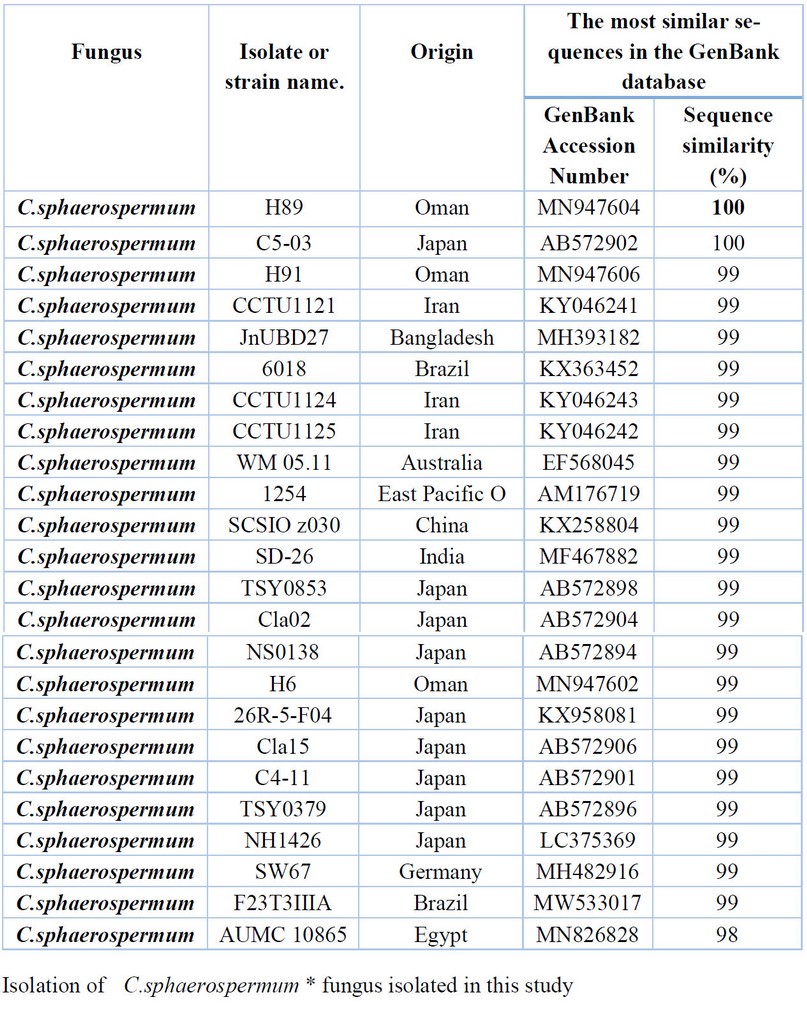
Table 10. Comparison between the similarity ratios of Cladosporium sphaerospermum isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
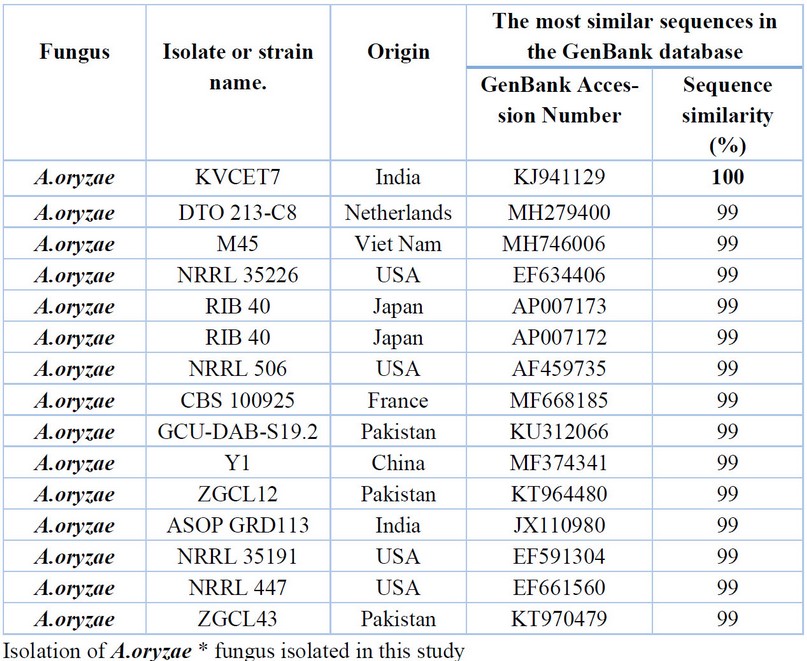
Table 11. Comparison between the similarity ratios of A.oryzae isolates from a wastewater treatment plant with the other isolate of the same fungi previously recorded in NCBI.
DISCUSSION
It shows that the highest percentage of 100% genetic similarity for A. caespitosus isolates (1,2,3,4,5,6) was with A.caespitosus isolates previously isolated from India and Thailand (MF319914 and KT385755, respectively), followed by the diagnosed fungal isolate in Slovakia (KM063244). India (HE653994), Kingdom of Saudi Arabia (MT328529), with a similarity rate of 99%, although genetically distant from the isolate of A. caespitosus isolated from Saudi Arabia (LN827684, with a difference of 97%. Also, It showed the proportion of similarity between this isolate and isolates from the world that has been recorded (NCBI). Furthermore, these isolates were not recorded in Iraq. Table 3. shows by comparing the DNA base the sequence of the dual-stranded DNA from A. flavus isolate (7,8) with the data at the (NCBI) that the highest similarity percentage (100%) was with the fungus isolates isolated from USA (CP051065, CP051089, CP051097) Uganda, India and China ( CP051033, MT292809 and CP047255), respectively. It was also found that the lowest genetic similarity percentage (99%) was for the fungal isolates isolated from India (MT509808, France, Italy, and Pakistan( MG745384, MN845204, KT970478). Respectively. These isolates from wastewater were not recorded in the Arab nations and Iraq.
The results presented in Table 4. show that the highest percentage of 100% genetic similarity for T.asperellum (9,10,11) isolates were with T.asperellum isolates previously isolated from Indonesia (LC514692) India (MK841023, MK253266), and the USA (MH130213) Isolates were recorded in the Arab world 100% similarity from Egypt (KC898194) and Morocco (MW074111). Although genetically distant from the isolate of T.asperellum isolated from India (MW493185) and Malaysia (MW082791) with a difference of 99%, The above Table Shows the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI). It was not recorded in the Arab nations and Iraq. The results presented in Table 5. were also presented by comparing the sequence of DNA's nitrogenous bases double for the A.tubingensis isolate (12) with data available in the (NCBI) that the highest genetic similarity rate of 100% with Previously isolated fungus isolates from China (MT446135, MT446097, MH055392) followed by the diagnosed fungus isolates from India (MT322427, MT443912) China (MT856267, MH055394), Nigeria (MH345877) and Mexico (MW193215) with a similarity rate of 99% The above Table It showed the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI). It was not recorded in the Arab nations and Iraq.
The results presented in Table 6. were also presented by comparing the sequence of the DNA's nitrogenous bases double for the A.terrus isolate (13) with data available in the (NCBI) that the highest genetic similarity rate of 100% with Previously isolated fungus isolates from China (KT310990) followed by the diagnosed fungus isolates from Iran (MN944451), South Korea (MN559622), China (KT310979, MK418744), Italy (MT316343), USA (NR_131276) and Egypt (MW282328) with a similarity rate of 99% The above Table It showed the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI), where it was not recorded in Iraq.
According to Table 7, comparing the series of the DNA's nitrogenous bases double for the A.alternate isolate (16) with the data available in the (NCBI) the highest genetic similarity rate of 100% with Previously isolated fungus isolates from South Korea (MK108918) followed by the diagnosed fungus isolates from India, Poland (MT524319, MW720803) China (MN589744, KJ002058, MW008923, KX0739949) and Iran (MG198619) with a similarity rate of 99% The above Table It showed the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI), where it was not recorded in the Arab nation and Iraq.
The results shown in Table 8 comparing the series nitrogenous bases of the DNA double A.niger isolates (14, 15) with the available Information in (NCBI) show that the highest genetic similarity rate of 100% with Previously isolated fungus isolates from China (KP764197), followed by the diagnosed fungus isolates in China (MT446087, MN429196) India (MN180811) with a similarity of 99% and also recorded in Egypt (LC582533)) with a similarity rate of 99%, The above Table It showed the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI), where it was not recorded in Iraq.
Table 9 shows that by comparing the DNA base and the sequence of the dual-stranded DNA from Acremonium sp. isolate (19) with the data at the (NCBI) only one isolate was recorded in the world, with a genetic similarity rate of 100%, with a fungus isolate previously isolated from Germany (AJ557731), where no recorded in the Arab world and Iraq.
The results presented in Table 10 were also presented by comparing the sequence of the DNA's nitrogenous bases double for the C.sphaerospermum isolate (17) with data available in the NCBI that a highest genetic similarity rate of 100% with Previously isolated fungus isolates from Oman (MN947604), Japan (AB572902) followed by the diagnosed fungus isolates from Oman (MN94760) Iran (KY046241, KY046243), Bangladesh (MH393182), Brazil (KX363452) and Egypt (MN826828) with a similarity rate of 99% The above Table It showed the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI).. Where it was not recorded in Iraq.
Table 11 are, comparing by the series of the DNA's nitrogenous bases double for the A.oryzae isolate (18) rate of 100% with Previously isolated fungus isolates from India (KJ941129) followed by the diagnosed fungus isolates from Netherlands (MH279400), Viet Nam (MH746006), USA (EF634406) Japan (AP007173) with a similarity rate of 99% The above Table It showed the proportion of similarity between this isolate and isolates from across the world that has been recorded (NCBI), where it was not recorded in the Arab nation and Iraq.
CONCLUSIONS
It is concluded from this study that there is an unlimited diversity of types of fungi in the wastewater treatment plant in Al-Barakia, Najaf. It is also concluded from this learning that all fungi isolates were genetically different from each other. The results proved that these isolates were previously registered in the center, as mentioned above. However, not all isolates were registered in Iraq. In this study, the technique of PCR was used in many previous studies to diagnose many organisms, including fungi, to get free of diagnostic harm depending on the morphological features of fungi 12.
Although morphological diagnosis helps to confine the fungi understudy into smaller sets before using other diagnostic ways, there are some problems associated with a morphological diagnosis of fungi as they require excellent experience in this area and a lot of time and effort. Since some fungal species are found very close to each other in some morphological features, the diagnostic process is more complicated 15. Moreover, fungi are affected by many environmental conditions that may alter the shapes, sizes, and colors of the spore and fungal colony19.
Differences in DNA sequences of the ITS region (internal transcription spacer) 20. They were highly effective and have been exploited to diagnose several fungi types. PCR-based identification has been used extensively in previous studies in diagnosing fungi such as Acremonium sp, Aspergillus flavus, Alternaria alternata, Cladosporium sphaerospermum, Aspergillus tubingensis, Aspergillus niger, 21,22,23
Although morphological diagnosis helps to confine the fungi understudy into smaller sets before using other diagnostic ways, there are some problems associated with a morphological diagnosis of fungi as they require excellent experience in this area and a lot of time and effort. Since some fungal species are found very close to each other in some morphological features, the diagnostic process is more complicated 15. Moreover, fungi are affected by many environmental conditions that may alter the shapes, sizes, and colors of the spore and fungal colony19.
Differences in DNA sequences of the ITS region (internal transcription spacer) 20. They were highly effective and have been exploited to diagnose several fungi types. PCR-based identification has been used extensively in previous studies in diagnosing fungi such as Acremonium sp, Aspergillus flavus, Alternaria alternata, Cladosporium sphaerospermum, Aspergillus tubingensis, Aspergillus niger, 21,22,23
Funding: "This research received no external funding."
Acknowledgments: I would like to express my sincere gratitude to Prof. Dr. Nihad Habeb Mutlak for his direct proposal on research and supervision and for giving me absolute confidence and guidance throughout this work. I extend my thanks and appreciation to Asst. Prof. Dr. Aqeel Nazal AL-Abedy for Financial and scientific assistance in completing this work
Conflicts of Interest: "The authors declare no conflict of interest."
REFERENCES
1. Metcalf & Eddy, Burton, F. L., Stensel, H. D., & Tchobanoglous, G. Wastewater engineering: treatment and reuse 2003.McGraw Hill.
2. Crini G. Recent developments in polysaccharide-based materials used as adsorbents in wastewater treatment. Progress in polymer science. 2005 Jan 1;30(1):38-70.
3. Cox M, Négré P, Yurramendi L. Industrial liquid effluents. INASMET Tecnalia, San Sebastian. 2007;283.
4. Hussien, S. A. .; Doosh, . K. S.. Extraction And Purification Of Β-Galactosidase From (Ziziphus Spina-Christi). JLSAR 2022, 3, 12-22.
5. Metcalf, Leonard, Harrison P. Eddy, and Georg Tchobanoglous. Wastewater engineering: treatment, disposal, and reuse 1991. Vol. 4. New York: McGraw-Hil,.
6. Krantz, D. and Kifferstein, B., Water pollution and society; 2005. Jungtinė Karalystė
7. .Henze M, Comeau Y. Wastewater characterization. Biological wastewater treatment: Principles modelling and design. 2008 Sep 1:33-52.
8. Kusters-van Someren MA, Samson RA, Visser J. The use of RFLP analysis in classification of the black Aspergilli: reinterpretation of the Aspergillus niger aggregate. Curr. Genet. 1991 Jan;19(1):21-6.
9. Gielkens M, Visser J, de Graaff L. Arabinoxylan degradation by fungi: characterization of the arabinoxylan-arabinofuranohydrolase encoding genes from Aspergillus niger and Aspergillus tubingensis. Curr. Genet.1997 Jan;31(1):22-9.
10. Hawksworth, D.L.; P.M. Kirk; B.C. Sutton and D.M. Pegler. Ainsworth & Bisby's dictionary of the fungi. 8th edition. CAB International Mycology Institute, Egham. 1995. C/589.203 H3
11. Ali, H.H., AL-Rawi, K., Khalaf, Y., Alaaraji, S., Aldahham, B., Awad, M., Al-ani, O., Al-ani, F., Ali, A.T.Serum Caveolin-1 Level is Inversely Associated with Serum Vaspin, Visfatin, and HbA1c in Newly Diagnosed Men with Type-2 Diabetes.Reports of Biochemistry and Molecular Biology,2022, 11 (2), pp. 299-309.
12. Khudai M Y, Abdulateef S M, Mouhammed T Th, Alamili H S. Use of modern geometric design of fish ponds to increase welfare and blood parameters. Revis Bionatura 2023;8 (2) 82. http://dx.doi.org/10.21931/RB/2023.08.02.82.
13. Symoens F, Bouchara JP, Heinemann S, Nolard N. Molecular typing of Aspergillus terreus isolates by random amplification of polymorphic DNA. J. Hosp. Infect. 2000 1 April;44(4):273-80.
14. Zuccaro A, Summerbell RC, Gams W, Schroers HJ, Mitchell JI. A new Acremonium species associated with Fucus spp., and its affinity with a phylogenetically distinct marine Emericellopsis clade. Stud Mycol. 2004 Jan 1;50:283-97.
15. Abdullah AA, Dewan MM, AL-Abedy AN. Genetic variation of some isolates of Cladosporium sphaerospermum isolated from different environments. In IOP Conference Series: Earth and Environmental Science 2019 1 November (Vol. 388, No. 1, p. 012016). IOP Publishing.
16. Zhang S, Zhao X, Wang Y, Li JI, Chen X, Wang A, Li J. Molecular detection of Fusarium oxysporum in the infected cucumber plants and soil. Pak. J. Bot. 2012;44(4):1445-51.
17. Prasad G, Kumar V, Dwivedi SK. Antifungal activity of some selected medicinal plants against Fusarium solani causing wilt and rot in Pearl millet. Asian J Bio Sci. 2018 Apr;13(1):21-7.
18. Hussain DF, Mutlag NH. Assessment the ability of Trichoderma harzianum Fungi in Bioremediation of some of Heavy Metals in Waste Water. In IOP Conference Series: Earth and Environmental Science 2021 1 June (Vol. 790, No. 1, p. 012087). IOP Publishing.
19. Yu JH, Butchko RA, Fernandes M, Keller NP, Leonard TJ, Adams TH. Conservation of structure and function of the aflatoxin regulatory geneaflR fromAspergillus nidulans andA. flavus. Curr. Genet. 1996 May;29(6):549-55.
20. A. Jassim, A., I. Al-Jugifi, W. Gradually Increasing In Lighting Intensity On Characteristics Of Ross 308 Broiler Carcass And Some Internal Organs. Anbar Journal Of Agricultural Sciences, 2023; 21(1): 205-213. doi: 10.32649/ajas.2023.179730.
21. Neamah SK, Alwan SL, AL-Abedy AN. Molecular diagnosis of new Aspergillus niger isolates causing degradation of pomegranate (Punica granatum) trees in Iraq. Plant Cell Biotechnol. Mol. Biol. 2020 Dec 31:79-84.
22. Torrance, L., Cowan, G. H., McLean, K., MacFarlane, S., Al-Abedy, A. N., Armstrong, M., ... & Bryan, G. J. (2020). Natural resistance to Potato virus Y in Solanum tuberosum Group Phureja. Theoretical and Applied Genetics, 133, 967-980.
23. Viterbo A, Montero M, Ramot O, Friesem D, Monte E, Llobell A, Chet I. Expression regulation of the endochitinase chit36 from Trichoderma asperellum (T. harzianum T-203). Curr. Genet. 2002 Nov;42(2):114-22.
Received: 26 September 2023 / Accepted: 15 April 2023 / Published:15 December 2023
Citation: Mutlag N.; Hussain D. New Records in Iraq and Arab Nations for some Fungi Isolated from Al-Barakia wastewater treatment plant in Al-Najaf Province. Revis Bionatura 2023;8 (4) 61. http://dx.doi.org/10.21931/RB/2023.08.04.61
Publisher's Note: Bionatura stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2023 by the authors. Submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
