2019.04.01.8
Files > Volume 4 > Vol 4 No 1 2019
INVESTIGATION / RESEARCH
Control of bacterial pathogens isolated from water using Actinomycetes extracts at Egerton University, Kenya
Paul Njenga Waithaka1, Eliud Mugu Gathuru2, Benson Muriuki Githaiga2, and Jackline Njeri Kamunyi2
Available from: http://dx.doi.org/10.21931/RB/2019.04.01.8
ABSTRACT
Diseases are the worst enemy to man currently. This study was aimed at isolating pathogenic bacteria from water obtained from shallow wells in Dundori Kenya. Also, the study aimed at testing the isolates for sensitivity to antibiotic metabolites previously extracted from Actinomycetes isolates from soils of Egerton University. Water samples were collected from shallow wells randomly selected from Dundori and abbreviated as A, B, C, D, and E. Bacterial pathogens were isolated from the water samples using the membrane filtration technique. The isolates were characterized using biochemical means. Antimicrobial sensitivity testing was carried out using Kirby Bauer disk diffusion method. Data analysis was carried out using the Statistical Package for Social Sciences (SPSS). Comparison of means was carried out using one way ANOVA. Shallow wells B, D and E were highly contaminated with pathogenic bacteria. Biochemical characterization of the isolates indicated that the most common isolates were Vibrio cholera, Klebsiella pneumoniae, Proteus sp, Escherichia coli, and Staphylococcus aureus. There was no significant difference between the zones of inhibition produced by the antibiotic metabolites (F=2.149 P=1340) when tested against the test isolates. There were no significant differences between the MIC's of the antibiotic metabolites on the bacterial pathogens (F=2.01 P=0.15). Water from some shallow wells in Dundori is highly contaminated with Klebsiella pneumoniae, Escherichia coli, Proteus sp., Vibrio cholerae and Staphylococcus aureus. The pathogens can effectively be controlled using antibiotics from the Actinomycetes. There is a need to sensitize the residents of the study area on ways of preventing seepage of contaminants into the shallow wells.
Keywords: Bacteria, Egerton, Control, Pathogens, Isolation, Water.
INTRODUCTION
Water from wells is considered contaminated or unsafe for drinking when tested for toxic chemicals or pathogenic microorganisms 1. The primary source of contamination by microorganisms could be fecal waste from warm-blooded animals including humans. Different pathogenic bacteria cause enteric diseases to humans while others are non-pathogenic 2. The water could also support the growth of protozoan and viral pathogens; hence water test for each pathogen would be extremely expensive and tedious 3 . Water is considered unsafe for human consumption when it has pathogenic microorganisms. Water test for bacteriological safety depends on microbiologists’ ability to detect coliform bacteria in the wells 4. Escherichia coli is the abundant bacteria in the test since it lives longer in water than other intestinal bacteria 5
Presence of pathogenic microorganisms in water sources may arise from contamination with fecal material, wastes from vegetation and animal dung resulting in growth of coliform bacteria 6. As a result, it becomes economically expensive for the communities to dig new wells with a low level of contamination 7. People who take contaminated water visit health facilities due to stomachache, diarrhea and flu-like symptoms. According to GOK (2014) 8 health statistics shows that pupils in Dundori have a high absenteeism rate and the cost of living in the area is rising at a very high rate.
In developing countries, contamination of water resources is indicated by the presence of enteric pathogens 9. However, rural communities use water disinfection methods such as chlorination, coagulation, sedimentation, and boiling 10. Some of these methods become impractical due to the high cost of the required equipment and low availability of chemical coagulants 11. Hundreds of people in Dundori area depend on both private and public wells which are infested with coliform bacteria. As a result, there is a need to control and improve the health safety of the water to levels that are considered safe for human consumption 12. The area is facing a challenge on how to offer more people access to clean and safe drinking water 13.
Actinomycetes have been used as potential sources of many bioactive compounds which have diverse clinical effects and essential applications in human medicine 14. Antibiotic metabolites screened from Actinomycetes isolated from soils of Egerton University may be a remedy to contamination of water with bacterial pathogens 15. The aims of this study were to isolate pathogenic bacteria from shallow wells of Dundori and carry out the sensitivity test of the isolates to antibiotics previously screened from Actinomycetes isolated from the soils of Egerton University.
MATERIALS AND METHODS
Study area
The study was carried out in Dundori ward with a population of 43,482. The geographical area is sloppy hence the risks of contamination of shallow wells with bacteria. According to Amata et al. (2014) 16, Dundori region has an average temperature of 17oC 63oF and lies at latitude 0.27 and longitude 36.1.
The Actinomycetes antibiotic metabolites
Four antibiotic metabolites coded EU30, EU37, EU41 and EU 154 had previously been extracted from Actinomycetes isolated from soils of Egerton University were used in this study. The Actinomycetes were grown in Luria Bertani broth for 7 days. The metabolites were extracted using ethyl acetate 17.
Water sampling
Five shallow wells coded A, B, C, D, and E were randomly selected from the study area. A total of 255 water samples were collected using sterile sample bottles. The sample size was determined using the formula by Waithaka et al. (2015) 18.

Where; n = sample size, p = anticipated prevalence which was 21% (0.21) in this study, q = failure which was calculated as 100-21 giving 79% (0.79), Z = is the appropriate value from the normal distribution for the desired confidence level which was 1.96 in this study, d = allowable error (0.05) and D = design effect. Based on a confidence interval of 95%, allowable error of 5% and a Z value of 1.96 the sample size was;
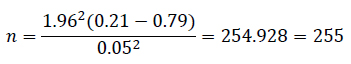
When collecting the samples, an empty ½ inch was left in the bottle for air space. The samples were transported to Egerton University Department of Biological Sciences Laboratories in iced containers and stored under refrigerated conditions at -4oC awaiting further processing.
Isolation of bacterial pathogens
One hundred milliliters water samples were filtered using membrane filters (Whatman GmbH, Germany), pore size 0.45μm, 47 mm diameter 19 . The membrane filters were aseptically placed on sterile nutrient agar in nutrient agar. Incubation was carried out at 37oC for 24h. Enumeration of bacteria was carried out with the help of a colony counter.
Biochemical characterization
Biochemical identification of the isolates was carried out using API 20E strip kit (bioMe´rieux®, Inc., France). The reagents used included API NaCl 0.85 % medium, API 20 E reagent kit, Zn reagent, oxidase, mineral oil, and API 20E Analytical Profile Index 20. The strips were prepared by the use of an incubation box (tray and lid). In this preparation, 5ml of distilled water was distributed into the honey-combed wells of the tray to create a humid atmosphere 21. Inocula of pure isolates were, emulsified in 5ml of normal saline 0.85% NaCl to achieve a homogeneous bacterial suspension. Anaerobiosis in the tests arginine dihydrolase (ADH), lysine decarboxylase (LDC), ornithine decarboxylase (ODC), H2S production (H2S), Urease (URE), citrate utilization (CIT), fermentation/oxidation of glucose (GLU), mannitol (MAN), inositol (INO), sorbitol (SOR), rhaminose (RHA), melibiose (MEL) and arabinose (ARA), gelatinase (GEL), tryptophane deaminase (TDA), fermentation/oxidation of sugars sucrose (SAC) and amygdaline (AMY), Indol (IND) and acetoin (VP) production was maintained by overlaying with mineral oil. The incubation box was closed and incubated at a temperature 37°C for 24h as described by the manufacturer, and the results were determined according to API 20E catalog. The microorganisms were further identified using colony morphology, Gram staining and motility test 22 .
Antimicrobial sensitivity testing
The antimicrobial susceptibility testing was carried out by use of Kirby Bauer disk diffusion method as described by the Clinical and Laboratory Standards Institute (CLSI) 22. Sterile wire loop was used to pick 3 colonies of each Klebsiella pneumoniae, Escherichia coli, Proteus Sp., Vibro cholera and Staphylococcus aureus. Emulsification in 3 ml of sterile physiological saline was carried out. Standardization of the suspended colonies was performed by diluting the normal saline suspension until the turbidity matched 0.5 McFarland standards. A sterile cotton swab was dipped into the standardized suspension, drained, and used for inoculating 20ml of Mueller-Hinton agar in sterile Petri plates. Discs (ABTEK BIOLOGICAL LTD., UK) 8mm in diameter were aseptically dipped into each of the antibiotic metabolites. The discs were air dried and placed on the agar using sterile forceps. The plates were incubated aerobically at 37°C for 24h. The zones of inhibition were measured in millimeters .
Statistical Analysis
Data obtained was presented in the form of tables. ANOVA was used to compare the means of viable cells in well A, B, C, D and E, zones of inhibition of the bacterial pathogens by the Actinomycetes crude extracts and minimum inhibitory concentration of the crude extracts.
RESULTS
Number of viable cells obtained from the shallow wells from Dundori
There were no viable cells isolated from well A and well C (Table 1). The number of viable cells isolated from well B varied from 25 to 46 per ml of water. However, in well D the range of viable isolated cells was 227 to 325. In well E, the viable cells ranged from 680 to 1020 per ml of water sample. There was a significant difference in the number of viable isolated cells in well A, B, C and D (F=154.265 P=0.0009).

Table 1: Number of viable cells (CFU/ml) isolated from well A, B, C, D, and E
Biochemical characteristics of the isolates
All the isolates apart from Vibrio cholarae were positive for catalase test. Also, the isolates were positive for the indole test except for Klebsiella pneumoniae and Proteus sp (Table 2). All the isolates were positive for Voges proskauer test except Escherichia coli and Proteus sp. However, the isolates were positive for citrate utelization except Escherichia coli and Vibrio cholerae.
On the other hand, all the isolates tested negative for methyl red except Proteus sp. and Vibrio cholerae. All the isolates tested negative for coagulase test except Staphylococcus aureus. Also, the isolates tested negative for oxidase test except for Staphylococcus aureus.

Table 2: Biochemical tests of the isolates
Zones inhibition of the isolates by the Actinomycetes metabolites
The zones of inhibition of K. pneumoniae varied from 21±0.2mm in EU 154 to 23±0.3mm in EU 30 (Table 3). In E. coli, the zones of inhibition ranged between 18±0.3mm in EU 37 to 23±0.2mm in EU 41. Besides, the zones of inhibition in Proteus sp. ranged between19±0.3mm in PAN 154 to 23±0.2mm in EU 30. However, in V. cholera the zones of inhibition varied from 18±0.2mm in EU 154 to 25±0.3mm in EU 41. In S. aureus, the zone of inhibition ranged between 22±0.3mm in EU 154 to 27±0.2 in EU 41. There was no significant difference between the zones of inhibition produced by the antibiotic metabolites (F=2.149 P=1340) when tested against the test isolates. The zones of inhibition were indicated by clearing around the disk having the antibiotic metabolites (Figure 1).

Table 3: Zones of inhibition of the isolated bacteria by metabolites from the selected Actinomycetes.
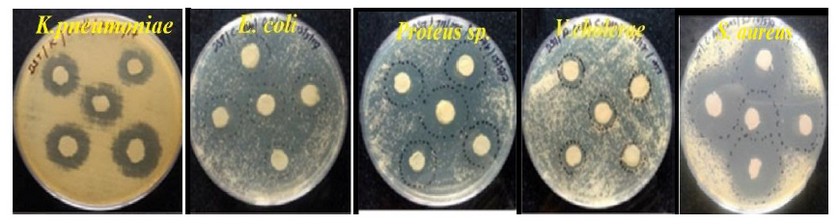
Figure 1: Zones of inhibition of the isolated bacteria by metabolites from the selected Actinomycetes.
Minimum inhibitory concentration (MIC) of the antibiotic metabolites
The minimum inhibitory concentration (MIC) in K. pneumoniae varied between 0.23±0.03mg/ ml in EU 41 to 0.25±0.01mg/ml in EU 30 (Table 4). In Escherichia coli, the MIC varied from 0.19±0.01mg/ml in EU 37 to 0.24±0.02mg/ml in EU 30. However, in Proteus sp. the MIC ranged between 0.18±0.01mg/ml in EU 41 and EU 154 to 0.25±0.02mg/ml EU 30. In V. cholera, the MIC varied from 0.17±0.02mg/ml in EU 41 to 0.25±0.02mg in EU 41. In addition, in S. aureus, the MIC varied from 0.23±0.01mg/ml in EU 41 to 0.25±0.01mg/ml in EU 30. There was no significant differences between the MIC’s of the antibiotic metabolites on the bacterial pathogens (F=2.01 P=0.15).
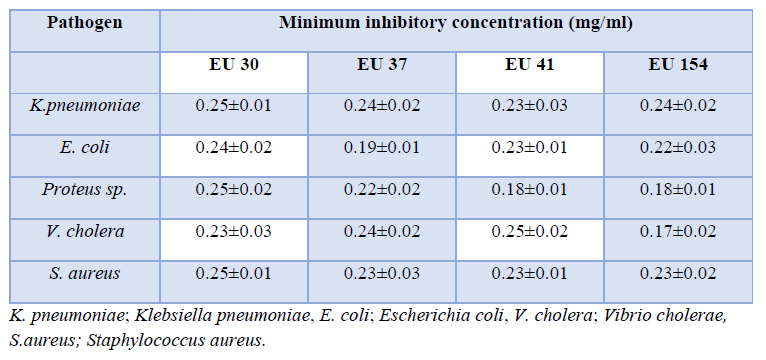
Table 4: Minimum inhibitory concentration of the isolated bacteria by metabolites from the selected Actinomycetes.
DISCUSSION
Shallow wells A and C had no viable cells (Table 1). However, viable cells were isolated from wells B, D and E. The lack of viable cells from wells A and C could be attributed to lack of seepage of wastewater in the area 18. The isolation of viable cells from wells B, D and E concurred with a study carried out in Pakistan 23 . The similarity in results between the two studies may be attributed to the proximity of the wells to pit latrines 24 .
The results on biochemical characterization of the isolates obtained in this study are typical of the isolates (Table 2). According to Joao and Cabra (2014) 25, biochemical characteristics of bacterial isolates are an indispensable tool in their identification. The results of the current study concur with those of a previous study by Poornima (2018) 26. The possible reason for the similarity in the results could be the isolation of the same bacteria 27. Florica et al. (2015) asserted that the same strains of bacteria react the same way biochemically.
The zones of inhibition of the bacterial pathogens by the Actinomycetes crude extracts are presented in Table 3. These results differed with a previous study obtained by Waithaka et al. (2017b) 15. Sukhvir et al. (2016) 28 explained that the type of antibiotics produced by Actinomycetes determines the zones of inhibition shown by the tested pathogens. Besides, the evolutionary characteristics of the test pathogens also determine the size of inhibition 29.
However, the minimum inhibitory concentrations presented by the metabolites in the current study partially agreed with a previous study by Rajeswari et al. (2015) 30. This may be attributed to the Actinomycetes utilizing the same mechanisms of inhibiting the growth of the test pathogens 31. In addition, the concentration of the antibiotics may have led to the observed results 32 .
CONCLUSIONS
Water from some shallow wells in Dundori is highly contaminated with K. pneumoniae, E. coli, Proteus sp., V. cholerae and S. aureus. The pathogens can effectively be controlled using antibiotics from the Actinomycetes.
RECOMMENDATIONS
There is a need to sensitize the residents of the study area on ways of preventing seepage of contaminants into the shallow wells. Further studies aimed at determining the mechanisms of antimicrobial control of actinomycete need to be carried out.
Acknowledgment
Thanks to the Department of Biological Sciences, Egerton University for giving us the laboratory space for carrying out this study.
Conflict of interest
No conflict of interest declared.
REFERENCES
1. Tao, R., Ying, G. G., Su, H. C., Zhou, H. W. and Sidhu, J. P. S. (2014). Detection of antibiotic resistance and tetracycline resistance genes in Enterobacteriaceae isolated from the Pearl Rivers in South China. Environmental Pollution; 158:2101-2109.
2. Lu, S. Y., Zhang, Y. L., Geng, S. N., Li, T. Y., Ye, Z. M., Zhang, D. S., Zou, F. and Zhou, H. W. (2014). High diversity of extended-spectrum beta-lactamase-producing bacteria in an urban river sediment habitat. Appllied Environmental Microbiology; 76:5972-5976.
3. Chen, Z., Yu, D., He, S., Ye, H., Zhang, L., Wen, Y., Zhang, W., Shu, L. and Chen, S. (2017). Prevalence of antibiotic-resistant Escherichia coli in drinking water sources in Hangzhou City”, Frontiers in Microbiology; 8:1133-1140.
4. David, W., Nageswara, R. and Narasimha, R. (2016). Concentration of organochlorine pesticide residues in sediments from the Godavari River of East Godavari District of Andhra Pradesh, Journal of Chemical, Biological and Physical Sciences; 3(3): 2279-2292.
5. Ateba, B. H., Nougang, M. E. and Noah, E. O. (2012). Occurrence of Salmonella in surface waters of Yaounde, Cameroon. Environmental Science and Water Research; 1: 243-50.
6. Dhanji, H., Murphy, N. M., Akhigbe, C., Doumith, M., Hope, R., Livermore, D. M., and Woodford, N. (2015). Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M- 14 extended-spectrum beta-lactamase from UK River water. Journal of Antimicrobial Chemotherapy; 66:512-516.
7. Abraham, W. R., Macedo, A. J., Gomes, L. H. and Tavares, F. C. A. (2016). Occurrence and resistance of pathogenic bacteria along the Tietê River downstream of São Paulo in Brazil. Braz. Journal of Biology; 35(4): 339-47.
8. Government of Kenya, (2014).Kenya National Bureau of Statistics, Kenya Population and Housing Census.Ministry of Planning and Development.Government printer, Nairobi, Kenya.
9. Goñi-Urriza, M., Capdepuy, M., Arpin, C., Raymond, N., Caumette, P., Quentin, C. (2014). Impact of an urban effluent on antibiotic resistance of riverine Enterobacteriaceae and Aeromonas spp. Applied and Environmental Microbiolology; 66:125-132.
10. Katakwar, M. (2016). Narmada river water: Pollution and its impact on the human health (2016). International Journal of Chemical Studies, 4(2):66-70.
11. Koike, S., Krapac, I. G., Oliver, H. D., Yannarell, A. C., Chee-Sanford, J. C., Aminov, R. I. and Mackie, R. I. (2015). Monitoring and Source Tracking of Tetracycline Resistance Genes in Lagoons and Groundwater Adjacent to Swine Production Facilities over a 3-Year Period. Applied Environmental Microbiology; 73:4813-4823.
12. Lupo, A., Coyne, S. and Berendonk, T. U. (2016). Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Frontiers in Microbiology; 3:18-39.
13. Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., Harbarth, S., Hindler, J. F., Kahlmeter, G., Olsson-Liljequist, B., Paterson, D. L., Rice, L. B., Stelling, J., Struelens, M. J., Vatopoulos, A., Weber, J. T. and Monnet, D. L. (2016). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clinical Microbiology and Infection; 18:268–281.
14. Panneerselvam, A. and Arumugam, G. (2015). Isolation and identification of bacteria from Lake water in and around Ranipet area, Vellore District. International Journal of Pharmaceutical and Biological Archives; 3(4):1008-1011.
15. Waithaka, P.N., Mwaura, F. B., Wagacha1, J. M., Gathuru, E. M. and Githaiga, B. M. (2017b). Antimicrobial properties of Actinomycetes isolated from Menengai crater in Kenya. CellBio; 6:13-26.
16. Amata, R. L., Otipa, M. J., Waiganjo, M., Wabule, M., Thuranira, E. G., Erbaugh, M. and Miller, S. (2014). Incidence, prevalence and severity of passion fruit fungal diseases in major production regions of Kenya. Journal of Applied Biosciences; 20:1146-1152.
17. Waithaka, P. N., Gathuru, E. M., Githaiga, B. M., Ochieng, E. O. and Linet, L.T. (2017a). Microbial Degradation of Polythene using Actinomycetes Isolated from Maize Rhizosphere, Forest and Waste Damping sites within Egerton University, Kenya. International Journal on Emerging Technologies; 8(1): 05-10.
18. Waithaka, P. N., Muthini, J. M. and Nyamache, A. K. (2015). Physico-chemical Analysis, Microbial Isolation, Sensitivity Test of the Isolates and Solar Disinfection of Water Running in Community Taps and River Kandutura in Nakuru North Sub-county, Kenya. The Open Microbiology Journal; 9: 117-124.
19. Rahube, T. O. and Yost, C. K. (2015). Antibiotic resistance plasmids in wastewater treatment plants and their possible dissemination into the environment. African Journal of Microbiology Research; 9:9183-9190.
20. Saghee, M. R. and Rajkumar, B. (2018). Organochlorine tolerant microbial populations profiled from Rivers Yamuna and Godavari using next generation sequencing. International Journal of Research in BioSciences; 7(1): 64-69.
21. Sharma S., Vishwakarma, R., Dixit, S. and Jain P. (2015). Evaluation of water quality of Narmada River with reference to physcochemical parameters at Hoshangabad city, MP, India”, Research Journal of Chemical Sciences; 1(3):123-130.
22. Atieno, R. N., Okemo, P. O. and Ombori, O. (2013). Isolation of high antibiotic resistant fecal bacteria indicators, Salmonella and Vibrio species from raw abattoirs sewage in Peri-urban locations of Nairobi, Kenya. Greener Journal of Biological Sciences; 3(5): 172-8.
23. Ehsan, H., Aqsa, B., Atif, U., Rehman, S. A. and Nodia, S. (2015). Isolation and identification of coliform bacteria from drinking water sources of Hazara Division, Pakistan. IOSR Journal of Pharmacy; (5)4:36-40.
24. Jain, D., Rahi, D. C. and Verma, S. (2017). Assessment of water quality status by using Water Quality Index (WQI) method in Narmada River in Jabalpur Madhya Pradesh. International Journal for Scientific Research & Development; 4 (12):122- 124.
25. João P. and Cabral, S. (2014). Water microbiology. Bacterial pathogens and water. International Journal of Environmental Research and Public Health, 7:3657-3703.
26. Poornima, S. (2018). Water quality of River Narmada at Gwari Ghat Jabalpur (India) in terms of microbial load, drug resistance and potability. Journal of Applied and Environmental Microbiology; 6(1):25-29.
27. Florica, M., Luminita M., Ioana, S. and Veronica L. (2015). Antibiotic resistance markers among Gram-negative isolates from wastewater and receiving rivers in South Romania. Romanian Biotechnological Letters; 20(1):10055-10069.
28. Sukhvir, K., Harjot, P. K. and Gagandeep, K. (2016) Isolation and Characterization of Antibiotic Producing Actinomycetes from Agriculture Soil. World Journal of Pharmaceutical Sciences; 5: 1109-1117.
29. Mobeen, S., Girija, S.G., Iswarya, M. and Rajitha. P. (2017). Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain Sankarensis-A10. Journal of Genetic Engineering and Biotechnology; 15, 87-94.
30. Rajeswari, P., Jose, P.A., Amiya, R. and Jebakumar, S.R.D. (2015) Characterization of Saltern Based Streptomyces sp. and Statistical Media Optimization for Its Improved Antibacterial Activity. Frontiers in Microbiology; 14:00753-00770.
31. Charousova, I., Medo, J., Halenarova, E. and Javorekova, S. (2017). Antimicrobial and Enzymatic Activity of Actinomycetes Isolated from Soils of Coastal Islands. Journal of Advanced Pharmaceutical Technology and Research; 8, 46-51.
32. Ahmed, I. K., Eltahir, H. B. and Humodi, A. S. (2016). Streptomyces: Isolation, Optimization of Culture Conditions and Extraction of Secondary Metabolites. International Current Pharmaceutical Journal; 5:27-32.
Received: 20 September 2018
Accepted: 12 January 2019
Paul Njenga Waithaka1, Eliud Mugu Gathuru2, Benson Muriuki Githaiga2 and Jackline Njeri Kamunyi2
1School of Biological Sciences, University of Nairobi, P. O. Box 30197 Nairobi, Kenya.
2Department of Biological Sciences, Egerton University, P. O. Box 536, Njoro, Kenya.
Corresponing author; Paul Njenga Waithaka, email; [email protected]
