CS 2019.02.01.24
Files > Conference Series > 2019 > Humboldt Kolleg 2019
Bionatura Conference Series Vol 2. No 1. 2019
“Breaking Paradigms: Towards a Multi-, Inter- and Transdisciplinary Science” In commemoration of the 250th Anniversary of Alexander von Humboldt
Bacteriophages applications in agriculture
Juan Carlos Laglaguano and Airina Valentina Cordova
available in: http://dx.doi.org/10.21931/RB/CS/2019.02.01.24
ABSTRACT
The bacteriophages life cycle has two stages: a lytic stage where the phages reproduce inside the bacteria and lyse bacteria and a lysogenic stage where the phage is in a stationary stage where do not exist phage reproduction. The understanding of the life cycle of phages is fundamental to understand the advantages of phage offers as biological control applications and how engineered phages work. The bacteriophages are an alternative to fight against the antimicrobial or pesticides because phages offer advantages such as high host specificity, the ability of long term effect, are active against dividing or not dividing bacterial cells, effective elimination of biofilms and are capable vehicles for nucleic acids delivery. Phages have been isolated from water or soil samples in different parts of the world and for specific bacterial pathogens. In the following review, in the main topics in bacteriophages and its applications in agriculture: the bacteriophages life cycle, advantages of phages in biological control applications, the last isolated phages and described for different pathogens and the last advances in phage engineering applications for biological control.
Keywords: bacteriophages, biological control, plant pathogen, phage therapy, agriculture.
INTRODUCTION
Agriculture as a traditional and economic activity around the world has been showing an increment of 1,0 % p.a in the present decade, in contrast to growth percentage (1,6% p.a) of 32 years ago. The estimations indicate a decline over time caused by several reasons such as change alimentary customs, global policies, territory, and change in environmental conditions.
Additionally, historical data reports that from 1950 the inclusion of antibiotics (for example Streptomycin, Gentamycin, so on) to cure the diseases caused by pathogenic bacteria in the crops1,2 in order to control the environmental factor that could cause a negative effect in the agriculture. However, the repetitive use of antimicrobial and antifungal compounds have generated that bacteria evolve developing a resistance. The resistance causes a decrease in efficacy to kill the pathogens for instance: Erwinia amylovora3 and Burkholderia pseudomalle4 bacterial pathogens and spread of diseases to humans because contaminated food5,6. Consequently, several biological fields have proposed bacteriophages as a plausible solution in this agricultural issue.
Phages are viruses that were discovered by two scientists: a pathologist named Frederick Twort at 1915 studying Micrococcus gram-positive bacteria and Félix d´Hérelle a microbiologist who found an “invisible anti microbe” in 19177,8. General terms, host bacterial cell, is infected by attachment of phage that transfers its genetic material through the cell membrane (or wall membrane in the case) and carrying out the genetic transcription, ending in the death of the host cell9. That evidence allow to bacteriophage serves like a biological control in the crops and agriculture problems, also known as phage therapy, which is a central topic of the present review.
Bacteriophages life cycle
The bacteriophages use different strategies to attack bacteria by introducing their DNA into the bacterias or by producing lysins. The bacterial life cycle of the bacteriophages there exists two phases: a lytic and a lysogenic stage.
Lytic cycle
During the lytic stage, the bacteriophages introduce their nucleic acid into the bacteria, by the action of an enzymatic mechanism for penetrating the peptidoglycan layer and inner membrane and ensure the release the phage DNA. In the bacteria cytoplasm, phage nucleic acids are susceptible to bacteria defense systems as restriction enzymes or exonucleases. For avoiding phage nucleic acids degradation, phages circularize their DNA by connecting their sticky ends, inhibit host nucleases, or add an odd nucleotide in their DNA. In some cases, phages genomes have evolved to eliminate sites that would be cut by the restriction enzymes of bacterial hosts. The assembly of phage elements such as capsid, phage nucleic acids packing, and tails occur in different stages within the bacteria organelles. The release of two enzymes performs the lysis of the bacteria, lysin, and holin. Lysins attack the peptidoglycans in the cell wall with the help of holin to reach the cell wall peptidoglycans and break the main bonds 10 ,11. Holin permeabilizes the membrane when achieving a specific concentration at which the cell membranes is lysed10.
Lysogenic cycle
The phage DNA will stay inside the bacteria without phage production in a quiescent state, a prophage. In the bacterial genome, the prophage genes are inserted near gal genes. Two genes are expressed to produce CI and Cro proteins. Both proteins compete to take the phages to lytic or lysogenic stages. CI gene activates a mechanism to bind operators to repress the expression of the rest of the phage genes. Two proteins, CII and CIII, promote the transition from lysogenic to the lytic stage11; these proteins bind promoters and stimulate transcription.
Antibiotic resistance
Over the decades, the use of substances against pathogens that damages plants and entire crops were common in the international range. Antibiotic resistance has been generating trouble in the agriculture sector due to what is currently known adverse effect: antibiotic resistance is even worst. Thus, study about Streptomycin resistance in E. amylovora bacterial pathogen, that is dangerous to apple and pears crops, has revealed two mechanisms by which one microorganism can resist to the antibiotic effect: Acquiring gen-resistance (pEA29 & pEa34) or through its chromosomal mutation2 which was confirmed by polymerase chain reaction (PCR) and gene sequencing. One of those mechanisms is known as horizontal gene transfer (HGT) that in simple words, is about the transference of external plasmids or transposons and integrons due to storage ability between neighboring microorganisms12. The second one is the genetic mutation, it usually generates the inactivation of antibiotic using self-resistance developed by pathogen such are degradation of antibiotic; changing the specificity for antimicrobial molecule; decrease antibiotic kinetic or taken it as a prisoner; the drug is a dangerous molecule for bacteria, and an expulsion mechanism is activated13. Due to the antimicrobial resistance, the use of phages has been put back into the scientific labs due to the specific life cycle and structure that allows the viruses to infect pathogenic bacteria that damage crops, and consequently affect other biotic components.
Bacteriophages specificity
The phage therapy is the use of phages to kill bacteria, in a broad and specific way. Each bacteriophage has a tail that specifically recognizes and binds receptors on the host bacterial cell surface and deliver the DNA into the bacterial cytoplasm. The phage tails are formed by receptor-binding proteins that would be tail fibers, tail spikes, or tail tips; those structures recognize host receptors such as lipopolysaccharides, teichoic acids, and porins14. Despite the great diversity and specificity between receptor-binding protein and bacterial receptor there exist phages that have shown the capability to infect a broad range of strains or genus, knowing this is important when using phages as a single phage or a phages cocktail. The great diversity of receptor-binding proteins and bacterial receptors make it challenging to predict the mechanisms of interaction between bacteriophages and bacteria15, so it is crucial to study an know the binding mechanisms between different bacteriophages and bacteria. The specificity of the bacteriophages makes it perfect for agriculture applications because phages only attack specific bacteria; this will be used to promote protective bacterial flora of plants16.
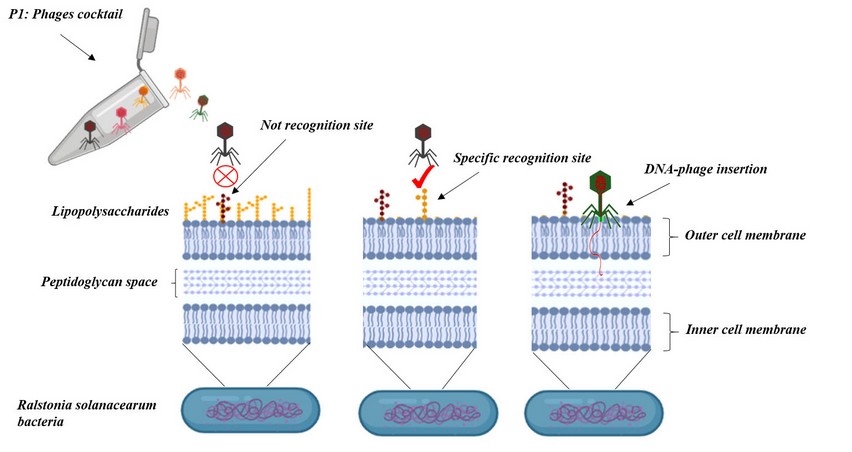
Fig 1. Bacteriophages specificity. Bacteriophages infect specific bacteria with specialized receptor molecules in the cell wall or membrane such as lipopolysaccharides, teichoic acids, and porins.
Phage enzymes
Phage lysins are hydrolytic enzymes that degrade cell wall peptidoglycans to help phage release. Lysins have been used successfully to eliminate Gram-positive and Gram-negative bacteria, having a tremendous rate of action and effectivity17. The lysins have been used against some resistant bacteria. Lysins also show a specific effect, due to act over specific peptidoglycans in the cell wall, the specificity would be species or genus, this characteristic is essential because lysins contribute to maintaining the bacterial flora of the plant and the soil15. Due to the characteristics mentioned before the engineering and discovery of new lysins by exchanging the domains of different lysins is a reality. Many of the engineered lysins have better antimicrobial activity than most of the antibiotics and never have been reported resistance against lysine18. Additionally, lysins can attack bacteria in biofilms, having a better performance than antibiotics or pesticides 15.
Last discoveries
During the last years, the efforts of discovery new phages or phage cocktails have been focused mainly in phages against Ralstonia solanacearum, which affect many plant species19. Last research in bacteriophages applied in agriculture is in Table 1.
Phage phiAP1
The phiAP1 is a phage of the family Podoviridae, has a close relation with the phage RSB3 another Ralstonia solanacearum phage. The phage phiAP1 was discovered in Brazil from soil samples from areas with incidence of bacterial wilt, specifically from the rhizospheric soil of symptomatic or asymptomatic plants. The phage was able to infect 50 of 60 different isolates of Ralstonia spp. collected from different disease, to survive desiccation or antibiosis in the soil and protect the bacteria from the plant defenses20. PhiAP1 phage contains two putative virion-associated peptidoglycan hydrolases (VAPGHs) that have not been reported in another Ralstonia spp. bacteriophages21. VAPGHs are phage-encoded lytic enzymes that degrade the bacterial peptidoglycan cell wall22. Some exciting genes found in the phiAP1 genomes were genes such as PagP-like gene, MarR gene, and Spanins genes. These genes are exciting because encodes for PagP-like gene encodes PagP enzymes that act as a helper of the lytic machinery, MarR gene encodes repressors of the transcription of some proteins of the outer membrane and spanins genes which encodes for molecules for host lysis during stress situations23.
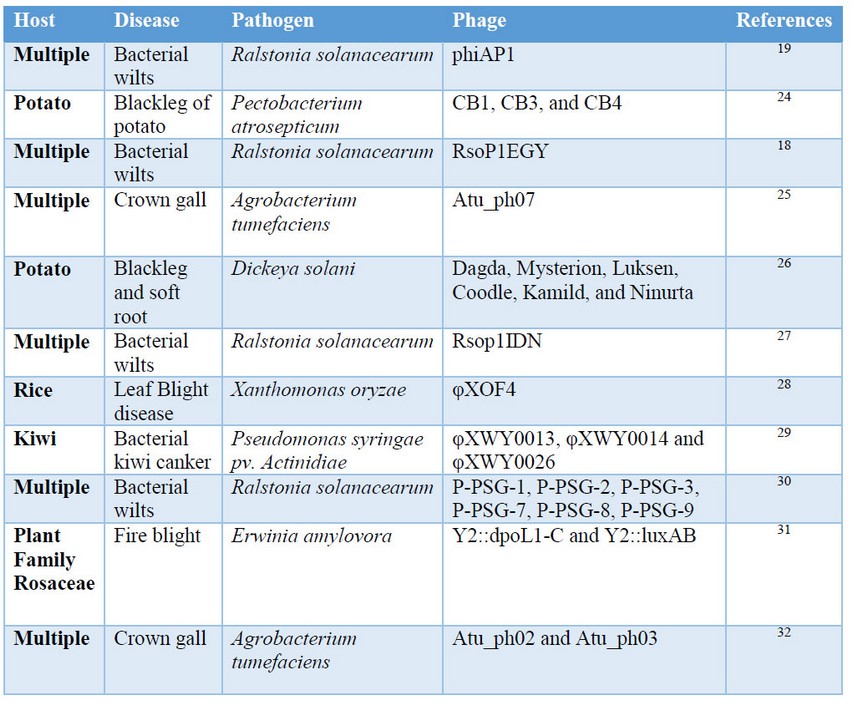
Table1. Bacteriophages discovered and tested for biological control applications in the last years.
CB1, CB3, and CB4 phages
The phages are from the virus family Podoviridae and have a close relationships with the N4 phage, an Escherichia phage. The phages were isolated of potato stem samples symptomatic for blackleg from Cork, Ireland. The phages CB1 and CB3 infect 12 of 19 exposed Pectobacterium atrosepticum strains while CB4 infect 10 of 19 strains, collectively can infect 15 of 19 different strains. The phages have a broad host spectrum in the Pectobacterium atrosepticum strains for biocontrol application however the phages do not infect another pathogen that causes soft root disease such as P. carotovorun, D. solani, and P. parmentieri. The phages will be used in a cocktail to treat soft root disease24.
RsPod1EGY phage
The phage was isolated from different soil samples of the potato rhizosphere from different locations around the Nile Delta in Egypt. The phage is specific for Egyptian R. solanacearum strains such as K3, K9, K10, K11, K12, K16, K17, and K19. The RsPod1EGY phage produces plaques with a diameter from 3.0 to 4.0 mm, which indicate a high lytic activity, which suggests the presence of depolymerase genes in the genome. The phage temperature range up is from 28 to 60 °C and a pH from 5 to 918.
Atu_ph07 phage
The Atu_ph07 phage was isolated from water samples from Sawyer Creek in Springfield. The genomic characterization of the phage suggests that is a lytic phage. The phage infected 8 of 9 strains of Agrobacterium tumefaciens biovar 1. The susceptible strains are LMG215, 06-777-2L, Chry5, LBA4404, A47a, C58, EHA 105, AGL-1, and GV3101. The phage contains genes encoding candidate lysis proteins (lysozymes) close to the head and tail phage, which will indicate those proteins are involved in the phage DNA entry25.
Six Dickeya solani phages
The phages were isolated from wastewater and organic waste from different locations in Denmark. It was isolated 46 bacteriophages that infect D. solani, according to their DNA homology, the phages were clustered into three groups with less than 0.2% DNA sequence similarity between groups and more than 48% similarity inside the groups. The three groups, obtaining at the end three groups named: cluster A, cluster B and cluster C. The analysis in BLAST suggested that just cluster A has new phages, not isolated or described previously. To develop a cocktail against D.solani were selected the following phages Dagda, Mysterion, Luksen, Coodle, Kamild, and Ninurta, selecting at least one phage of each cluster to avoid that the host develops resistance. The treatment with the cocktail shows a significant reduction in the infection26.
RsoP1IDN phage
The phage PsoP1IDN was isolated from soil samples of an eggplant field in Jember, Indonesia. The phage is of the Pradovirus genus and close related to RSB1 phage. The phage was tested in 16 strains of R. Solanacerum from different parts of the world, being susceptible for three R. Solanacearum strains: K60, RUN302, and DT3. The experiments showed a plate formation of approximately 9.15 ± 1.63 in diameter. The genome analysis suggests that is a lytic phage due to the presence of antimicrobial proteins, such as lysozymes33.
φXOF4 phage
The phage φXOF4 was isolated from soil and water samples collected Andhra Pradesh in India, a region considered as a hotspot of leaf blight disease. The phage is of the family Siphoviridae, order Caudovirales. Eighth phages were isolated and tested in 5 strains of Xanthomonas oryzae: PPXAP1, PPXAP2, PPXAP3, PPXAP4, and PPXAP5. The phage φXOF4 was able to infect the 6 tested Xanthomonas oryzae strains. In the spot test experiments the phage φXOF4 formed a plate from 3 to 4 mm of diameter. The phage show to be useful to disinfect seeds, eliminating the presence of Xanthomonas oryzae28
φXWY0013, φXWY0014, and φXWY0026 phages
The phages were isolated from superficial water samples around Shangai, China. A total of 36 phages of different families: Podoviridae, Siphoviridae, and Myovridae were isolated; however, just three phages, one of each family were selected for the study. The three selected phages show to be able to infect 14 of 52 strains of Pseudomonas syringae pv. strains isolated from different parts of China and all the infected strains were of the biovar 3. The temperature range of infectivity of the phages is from 25 to 60°C and are inactivated at 70°C. The optimum phage pH is from 2.0 to 12. The phylogenetic studies of the phages suggest that are lytic phages29.
P1 phage cocktail (P-PSG-1, P-PSG-2, P-PSG-3, P-PSG-7, P-PSG-8, and P-PSG-9)
The phages were isolated from different water sources in China and Kenya. From 18 water samples, only 12 bacteriophages show antimicrobial activity against the tested R. solanacearum strains. The Ralstonia Solanacearum strains tested in the experiments were PS-X4-1, PS-X10-2, and PS-13-1. The phages were tested in their capability to kill bacteria in the time, R. solanacearum strains were irradiated with different phages. Each hour was evaluated their optical density. The samples treated with phages that show a decreasing of the optical density in the time that means a decreasing in the growing of R. solanacearum. The bacteriophages that show the higher decreasing in the bacteria growth were used in the development of the P1 phage cocktail30.
Atu_ph02 and Atu_ph03 phages
The phages were isolated from water samples of the Columbia regional wastewater treatment plant in EEUU. The A. tumefaciens strains were isolated from cherry tree tumors. The tested strains were C58, EHA105, EHA101, GV3101, NTL4, and AGLL-1. All the strains were susceptibles to both phages except the strain AGLL-1. AGLL-1 strain has a mutation in the gene RecA, a gene responsible for homologous recombination and DNA repair. The observations suggest that the RecA gene is essential for efficient infection. In silico analysis of the phages, shows the presence of 2 peptid-glycogen hydrolases, which will be involved in the bacteria lysis32.
Engineered phages
The understanding of bacteriophages biology and the development of new engineering tools to modify DNA as CRISPR-Cas allow the construction of new strategies to eliminate pathogens34,35. The capability of bacteriophage to introduce nucleic acids have been exploited to construct new strategies such as introduce genes to produce bacteriostasis, synthesis of harmful compounds or insert genes to restore the resistance against antibiotics15. For example, Pseudomonas aeruginosa a pathogen that causes soft root was modified by the exchange of lysin genes to restriction enzyme genes to produce bacteriostasis17. The strategy to eliminate resistance is to introduce genes to inhibit resistance genes36. In the case of induction to produce harmful compounds, are introduced genes of toxins as Gef and ChpBK. Gef and ChpBK proteins are toxic proteins derived from the toxin-antitoxin system. Gef interferes with the toxic-antitoxic system and depolarizes the membrane while ChpBK cleaves mRNA, which inhibits protein translation37.
Engineered phage Y2::luxAB
The luxAB phage was created by the introduction of the luxAB gene by homologous recombination during infection of E. amylovora into the phage Y2. The luxAB gene encodes for a luciferase. The idea of introducing a luciferase gene is to use it as a bioreporter of the infected host bacteria. The Y2::luxAB phage introduce luciferase gene into the bacteria, then the bacteria can produce luciferase when it is infected. The introduction of the luxAB gene does not affect the lytic function of phage. The reporter works; however, the luminescence signal decreases rapidly, probably due to the bacteria lysis caused by the phage31.
Engineered phage Y2::luxAB
The dpoL1-C gene, a depolymerase gene was introduced by homologous recombination during infection of E. amylovora into the Y2 phage. The introduction of the dpoL1-C gene increment the lytic function of the phages compared with the parental phage; this was evaluated by measuring the size of the phage plate and the infected bacteria after 24h. The bacteria infected with Y2::luxAB phage present a higher phage plate size and a higher number of infected bacteria after 24h31.
Taking advantage of the specificity of bacteriophages
The development of genetic engineering techniques allows creating new strategies to take advantage of phages specificity. Saccharomyces cerevisiae is a model organism able to easy modification was used to clone the phage genome. Multiple fragments of T3 and T7 phages genome were amplified by PCR and assembled leaving ends that have homology with the yeast artificial genome (YAC). During the assembly, for example, E.coli infecting scaffolds were introduced into another pathogenic bacteria Klebsiella and Yersinia. The phage genome and YAC are ensembled, transformed, and cloned inside the yeast, creating a YAC-phage genome. The introduced scaffolds allow amplifying the hosts' target of phage and keep the phage specificity38.
REFERENCES
1. Tancos KA, Borejsza-Wysocka E, Kuehne S, Breth D, Cox KD. Fire Blight Symptomatic Shoots and the Presence of Erwinia amylovora in Asymptomatic Apple Budwood. Plant Dis. 2016 Aug 29;101(1):186–91.
2. McManus PS, Stockwell VO, Sundin GW, Jones AL. Antibiotic use in plant agriculture. Annu Rev Phytopathol. 2002;40:443–65.
3. Koskella B, Taylor TB. Multifaceted Impacts of Bacteriophages in the Plant Microbiome. Annu Rev Phytopathol. 2018 25;56:361–80.
4. Guang-Han O, Leang-Chung C, Vellasamy KM, Mariappan V, Li-Yen C, Vadivelu J. Experimental Phage Therapy for Burkholderia pseudomallei Infection. PLOS ONE. 2016 Jul 7;11(7):e0158213.
5. Chang Q, Wang W, Regev-Yochay G, Lipsitch M, Hanage WP. Antibiotics in agriculture and the risk to human health: how worried should we be? Evol Appl. 2015 Mar;8(3):240–7.
6. Thanner S, Drissner D, Walsh F. Antimicrobial Resistance in Agriculture. mBio. 2016 May 4;7(2):e02227-15.
7. Afazeli H, Jafari A, Rafiee S, Nosrati M. An investigation of biogas production potential from livestock and slaughterhouse wastes. Renew Sustain Energy Rev. 2014 Jun 1;34:380–6.
8. On an invisible microbe antagonistic to dysentery bacilli. Note by M. F. d’Herelle, presented by M. Roux. Comptes Rendus Academie des Sciences 1917; 165:373–5: Bacteriophage: Vol 1, No 1 [Internet]. [cited 2019 May 7]. Available from: https://www.tandfonline.com/doi/abs/10.4161/bact.1.1.14941
9. Basdew IH, Laing MD. Mini-Review: Biological control of bovine mastitis using bacteriophage therapy. In 2011.
10. Wang I-N, Deaton J, Young R. Sizing the holin lesion with an endolysin-beta-galactosidase fusion. J Bacteriol. 2003 Feb;185(3):779–87.
11. Fischetti VA. Bacteriophage lytic enzymes: novel anti-infectives. Trends Microbiol. 2005 Jan 1;13(10):491–6.
12. Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014 Nov;32(11):1146–50.
13. Hagens S, Habel A, Ahsen U von, Gabain A von, Bläsi U. Therapy of Experimental Pseudomonas Infections with a Nonreplicating Genetically Modified Phage. Antimicrob Agents Chemother. 2004 Oct 1;48(10):3817–22.
14. Yap ML, Rossmann MG. Structure and function of bacteriophage T4. Future Microbiol. 2014;9(12):1319–27.
15. Lin DM, Koskella B, Lin HC. Phage therapy: An alternative to antibiotics in the age of multi-drug resistance. World J Gastrointest Pharmacol Ther. 2017 Aug 6;8(3):162–73.
16. Gray JA, Chandry PS, Kaur M, Kocharunchitt C, Bowman JP, Fox EM. Novel Biocontrol Methods for Listeria monocytogenes Biofilms in Food Production Facilities. Front Microbiol [Internet]. 2018 [cited 2019 May 7];9. Available from: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00605/full
17. Schmelcher M, Donovan DM, Loessner MJ. Bacteriophage endolysins as novel antimicrobials. Future Microbiol. 2012 Oct;7(10):1147–71.
18. Ahmad AA, Elhalag KM, Addy HS, Nasr-Eldin MA, Hussien AS, Huang Q. Sequencing, genome analysis and host range of a novel Ralstonia phage, RsoP1EGY, isolated in Egypt. Arch Virol. 2018;163(8):2271–4.
19. Morel A, Peeters N, Vailleau F, Barberis P, Jiang G, Berthomé R, et al. Plant Pathogenicity Phenotyping of Ralstonia solanacearum Strains. In: Medina C, López-Baena FJ, editors. Host-Pathogen Interactions: Methods and Protocols [Internet]. New York, NY: Springer New York; 2018 [cited 2019 May 7]. p. 223–39. (Methods in Molecular Biology). Available from: https://doi.org/10.1007/978-1-4939-7604-1_18
20. Involvement of Bacterial Polysaccharides in Plant Pathogenesis | Annual Review of Phytopathology [Internet]. [cited 2019 May 13]. Available from: https://www.annualreviews.org/doi/abs/10.1146/annurev.py.33.090195.001133
21. Kawasaki T, Narulita E, Matsunami M, Ishikawa H, Shimizu M, Fujie M, et al. Genomic diversity of large-plaque-forming podoviruses infecting the phytopathogen Ralstonia solanacearum. Virology. 2016 May;492:73–81.
22. Bacteriophage virion-associated peptidoglycan hydrolases: potential new enzybiotics. - PubMed - NCBI [Internet]. [cited 2019 May 13]. Available from: https://www.ncbi.nlm.nih.gov/pubmed/22991936
23. da Silva Xavier A, da Silva FP, Vidigal PMP, Lima TTM, de Souza FO, Alfenas-Zerbini P. Genomic and biological characterization of a new member of the genus Phikmvvirus infecting phytopathogenic Ralstonia bacteria. Arch Virol. 2018 Dec;163(12):3275–90.
24. Buttimer C, Hendrix H, Lucid A, Neve H, Noben J-P, Franz C, et al. Novel N4-Like Bacteriophages of Pectobacterium atrosepticum. Pharm Basel Switz. 2018 May 14;11(2).
25. Attai H, Boon M, Phillips K, Noben J-P, Lavigne R, Brown PJB. Larger than life: Isolation and genomic characterization of a jumbo phage that infects the bacterial plant pathogen, Agrobacterium tumefaciens. Front Microbiol. 2018;9(AUG).
26. Carstens AB, Djurhuus AM, Kot W, Jacobs-Sera D, Hatfull GF, Hansen LH. Unlocking the Potential of 46 New Bacteriophages for Biocontrol of Dickeya Solani. Viruses. 2018;10(11).
27. Addy HS, Farid MM, Ahmad AA, Huang Q. Host range and molecular characterization of a lytic Pradovirus-like Ralstonia phage RsoP1IDN isolated from Indonesia. Arch Virol. 2018 Dec;163(12):3409–14.
28. Ranjani P, Gowthami Y, Gnanamanickam SS, Palani P. Bacteriophages: A new weapon for the control of bacterial blight disease in rice caused by Xanthomonas oryzae. Microbiol Biotechnol Lett. 2018;46(4):346–59.
29. Yin Y, Ni P, Deng B, Wang S, Xu W, Wang D. Isolation and characterisation of phages against Pseudomonas syringae pv. actinidiae. Acta Agric Scand Sect B Soil Plant Sci. 2019;69(3):199–208.
30. Wei C, Liu J, Maina AN, Mwaura FB, Yu J, Yan C, et al. Developing a bacteriophage cocktail for biocontrol of potato bacterial wilt. Virol Sin. 2017;32(6):476–84.
31. Born Y, Fieseler L, Thöny V, Leimer N, Duffy B, Loessner MJ. Engineering of bacteriophages Y2::dpoL1-C and Y2::luxAB for efficient control and rapid detection of the fire blight pathogen, Erwinia amylovora. Appl Environ Microbiol. 2017;83(12).
32. Attai H, Rimbey J, Smith GP, Brown PJB. Expression of a peptidoglycan hydrolase from lytic bacteriophages Atu_ph02 and Atu_ph03 triggers lysis of Agrobacterium tumefaciens. Appl Environ Microbiol. 2017;83(23).
33. Addy HS, Farid MM, Ahmad AA, Huang Q. Host range and molecular characterization of a lytic Pradovirus-like Ralstonia phage RsoP1IDN isolated from Indonesia. Arch Virol. 2018 Dec 1;163(12):3409–14.
34. Schwarz H, Riede I, Sonntag I, Henning U. Degrees of relatedness of T-even type E. coli phages using different or the same receptors and topology of serologically cross-reacting sites. EMBO J. 1983;2(3):375–80.
35. Ingham SC, Losinski JA, Andrews MP, Breuer JE, Breuer JR, Wood TM, et al. Escherichia coli Contamination of Vegetables Grown in Soils Fertilized with Noncomposted Bovine Manure: Garden-Scale Studies. Appl Environ Microbiol. 2004 Nov;70(11):6420–7.
36. Endersen L, O’Mahony J, Hill C, Ross RP, McAuliffe O, Coffey A. Phage therapy in the food industry. Annu Rev Food Sci Technol. 2014;5:327–49.
37. Loureiro A, Da Silva GJ. Crispr-cas: Converting a bacterial defence mechanism into a state-of-the-art genetic manipulation tool. Antibiotics. 2019;8(1).
38. Goren MG, Yosef I, Qimron U. Programming Bacteriophages by Swapping Their Specificity Determinants. Trends Microbiol. 2015 Dec;23(12):744–6.
Received: 17 April 2019
Accepted: 23 May 2019
Airina Valentina Cordova
Afiliación: School of Chemical Sciences and Engineering. Yachay Tech University, San Miguel de Urcuquí, Ecuador
mail:[email protected]
https://orcid.org/0000-0003-4744-4103
Juan Carlos Laglaguano
https://orcid.org/0000-0003-0740-7113
afiliación: School of Biological Sciences and Engineering, Yachay Tech University, San Miguel de Urcuquí, Ecuador
mail: [email protected]
