2024..01.01.36
Files > Conference Series > 2024 > Chimboazo ild pagina nueva
Morphogenetic identification of new endophytic fungal species related to the family of Plectosphaerellaceae in Iraq
Alaa Alauldeen Al-Rifaie1* and Mohanad Khalaf Mohammed-Ameen1
Department of Biology, College of Science, University of Basrah, 61004, Basrah, IRAQ.
*Corresponding author's email: [email protected]
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.36
ABSTRACT
This study focused on isolating and identifying endophytic fungi from vegetable crops in Iraq. Samples from seven vegetable plants, including Anethum graveolens, Apium graveolens, Capsicum annuum, Malva parviflora, Mentha piperita, Petroselinum sativum, Portulaca oleracea, were collected from five central regions in Basrah, Iraq, (Abu Al-Khaseeb, Karmat Ali, AL-Zubair, Shatt Al-Arab and the Centre of Basrah). Samples, including mature leaves, stems and roots of vegetable sources, were collected and treated in the laboratory. Recovered endophytic fungi were purified and identified based on their macro and micromorphological features. Identification was validated by DNA sequencing and PCR amplification of ITS4 and ITS5 gene primers and molecular analysis. Phylogenetic examination indicated that three novel endophytic fungal species are documented in the Iraqi mycobiota for the first time, was isolated from vegetable plants in Basrah province related to the family Plectosphaerellaceae, including Gibellulopsis serrae, Plectosphaerella niemeijerarum, P. oratosquillae. Brief descriptions and photo panels are provided for the newly recorded species in this study. These findings are essential to understanding the endophytic fungal community within vegetable parts that can be used to manage and control plant disease and enhance productivity.
Keywords: Ascomycota, Bsarah, endophytic fungi, Iraq, Plectosphaerellaceae.
INTRODUCTION
Endophytic Fungi (EF) inhabit all healthy plant tissues throughout at least a portion of the plant life cycle without causing disease or noticeable morphological alterations 1. The presence of fungi within plant tissues has been recognized since the late 19th century, and the term "endophyte" was initially introduced by de Bary in 1866. An endophytic fungus lives in mycelial form in biological association with the living plant. EF is found in various plants, i.e., trees, grasses, algae and herbaceous plants.
Endophyte and endophytic fungi have been frequently used to describe the internal mycota of living plants. EF is now considered an essential component of biodiversity 2. Endophytic fungi are one of the most creative groups of secondary metabolite producers that play essential biological roles in human life. They are potential sources of novel natural agents for exploitation in the pharmaceutical industry, agriculture, and environmental applications. It also considers their medicinal applications, especially in producing anticancer, antimicrobial, antioxidant, and antiviral compounds. (EF) are highly diverse, with the reported majority being ascomycetes, and also lack a teleomorphic state 3. In one survey, it is estimated that over one million fungal endophytes exist in nature 4.
The family Plectosphaerellaceae (Glomerellales, Sordariomycetes, Ascomycota) was proposed based on the plant pathogen Plectosphaerella cucumerina as the type species 5. This family is distributed in various habitats and has wide geographic distribution, inhabiting different substrates, including plants, soil, and insects. Most species are pathogens, causing significant losses in agriculture 6. Currently, 10 genera are accepted in the family, i.e., Acrostalagmus, Brunneomyces, Chordomyces, Gibellulopsis, Lectera, Musicillium, Plectosphaerella, Sodiomyces, Stachylidium and Verticillium s. str. However, Cephalosporium serrae, Gliocladium cibotii and several Acremonium species are included in the family 7.
Some species, such as fruit, tomato, pepper, bamboo, and asparagus, are pathogens on plants 8. Plectosphaerella cucumerina is the most frequent pathogen of several plant species, causing fruit, root and collar rot and collapse. It is considered a severe threat to melon (Cucumis melo) production in Italy. They are causing significant losses of melon, pumpkin and zucchini crops 9. Besides diseased plants, some kinds of Plectosphaerella endophyte were reported as endophytic Plectosphaerella sinensis without causing apparent symptoms 10.
Most species of Plectosphaerella isolated from soil and plants except P. oratosquillae isolated from mantis shrimp (Oratosquilla oratorio) in Yamaguchi and Aichi Prefectures, Japan, are described as the new species Plectosporium oratosquillae, infect the gills of the mantis shrimp, which become brown or black due to melanization 11. The asexual morphs are more abundant in this family; few sexual morphs have been reported, and they have simple or verticillate conidiophores with phialidic conidiogenous cells and mostly cylindrical or ellipsoidal conidia arranged in slimy heads. Plectosphaerella cucumerina produces perithecial ascomata with clavate asci and hyaline, two-celled ascospores 7. In Iraq, very few studies focus on isolating and identifying endophytic fungi in Iraq 12,13. Some fungi, such as Fusarium and Aspergillus species 14–16, are considered severe plant pathogens.
Due to rare studies about the isolation and identification of the Plectosphaerellaceae family as endophytic fungi in Iraq and the Arabic world, this work was designed to investigate the fungal biodiversity of Plectosphaerellaceae family from vegetable resources via morphological characterization and using molecular sequencing validation to understand the existence and diversity of this family as an endophyte in Iraq. Therefore, the study is the first to report on many kinds of Plectosphaerella in Iraq and the Arabic world.
MATERIALS AND METHODS
Samples collection
During this study,36 plant samples were collected from five Basrah province, Iraq regions. (Abu Al-Kaseib, Karmat Ali, Al-zubair, ShattAl-arab and the Centre of Basrah) As shown in (Figure 1) between 01.01.2022 to 01.09.2022. Plant samples, including mature leaves, stems and roots, from 1/1/2022 to 1/9/2022. Samples are taken from different parts of plants, such as mature leaves, stems and roots. The samples were brought in clean plastic bags to the fungi laboratory of Basrah University - College of Science- Department of Life Sciences for sample handling and isolation process.
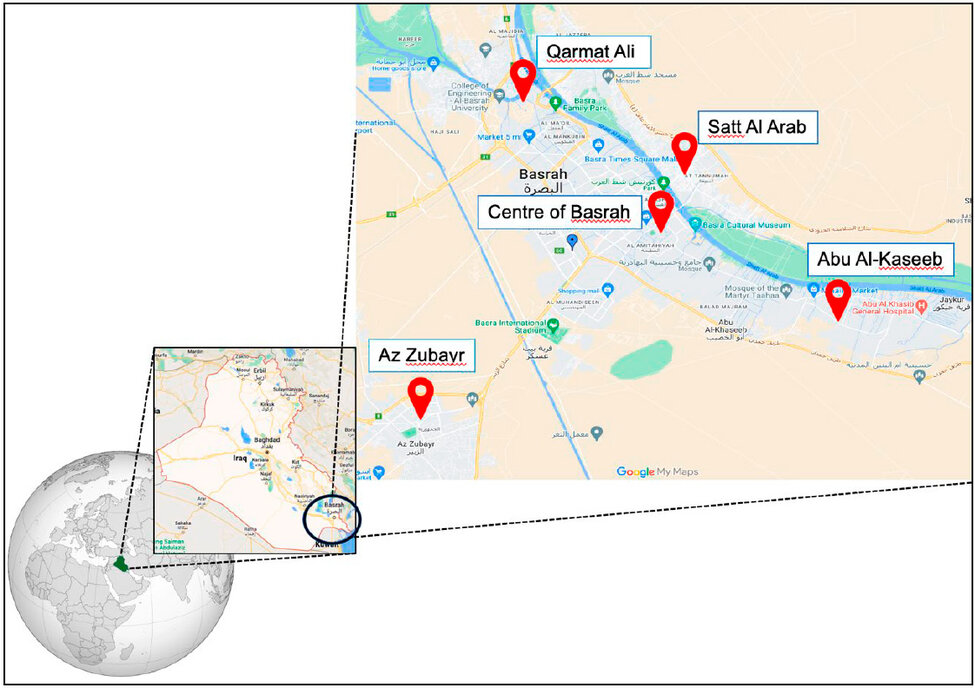
Figure 1: Location of vegetable samples collection for endophytic fungi isolation Basrah Province, Iraq.
Isolation and purification of endophytic fungi
Isolation fungal endophytes were carried on following two methods:
A. Solid culture media
After the surface layer of each sample was sterilized, they were thoroughly washed under running tap water to eliminate dust and debris. Next, the samples were washed with autoclaved distilled water. Vegetable samples were cut into small pieces with 2-4 mm length dimensions using sterile razor blades. To sterilize vegetable segments surfaces, a sequential immersion process was conducted with 70% ethanol for 60 seconds, 0.5% sodium hypochlorite for 5 minutes, 70% ethanol for 30 seconds, and then given a final rinse for 5 minutes with sterilized distilled water, and then, were air-dried using sterilized filter paper 12. Finally, vegetable segments were implanted on a Petri dish of Potato Dextrose Agar (PDA) supplemented with 0.01% Chloramphenicol antibiotic, and 3 replicates were made per plant part. The plates were carefully sealed and incubated at 25 °C for 5 – 7 days. Samples were monitored daily for the detection of the growth of endophytic fungi. Recovered fungi were picked and transferred into a new PDA plate for preservation and identification. The pure isolates of endophytic fungi were then separately transferred to PDA slants and a sterilized glycerol solution at a 50% (v/v) concentration for long-term preservation.
B. Moist chamber method
Vegetable samples were cut, measuring approximately 4 - 5 mm, then washed twice with tap water, followed by the same washing and sterilization process described above. The dried plant pieces were arranged in large Petri dishes (15 cm in diameter) containing sterile filter paper moistened with sterile distilled water to provide moisture. Subsequently, the Petri dishes were placed in an incubator at 25 ± 2 °C for 15 to 20 days. Plates were monitored daily for detection of the growth of endophytic fungi. Axenic cultures of isolated endophytic fungi were preserved in The Postgraduate Fungal Research Laboratory, Department of Biology, College of Science, University of Basrah, IRAQ.
Molecular identification of isolated endophytic fungi
Deoxyribonucleic Acid (DNA) of isolated endophytic fungi was extracted using (Yeast Genomic DNA Kit Cat. No. GBYB100, Geneaid, Taiwan). Briefly, the axenic culture of each endophytic fungi isolates on PDA was freshly sub-cultured on PDA and incubated for 7 days at 25 ± 2°C. Pure fungal biomass 50-200 mg was transferred to a clean 1.5 ml microcentrifuge tube for DNA extraction following the manufacturer's instructions. Extracted DNA of endophytic fungi was amplified by the ITS1-5.8S-ITS2 region sequencing of the rRNA encoding gene unit using the primers ITS5ext (5’-GTA ACA AGG TTT CCG TAG GTG-3') and ITS4 ext (5'TTC TTT TCC TCC GCT TAT TGA TAT GC3') 14. PCR amplification condition of ITS region was conducted 15. The amplified DNA bands of the fungal species were sent to Macrogen in South Korea for additional sequencing analysis. The species identity was determined based on the sequence identity of over 95% of the ITS region of recovered species using the Basic Local Alignment Search Tool (BLAST). It was compared with the GenBank sequence database at the National Centre for Biotechnology Information (NCBI). Alignments for each data set were made in (MEGA11) 16. Sequence phylogeny was generated using Maximum Likelihood (ML) with the best nucleotide substitution model and the Jukes-Cantor model with the lowest BIC scores (Bayesian Information Criterion). The bootstrap consensus tree was inferred from 1000 replicates. After genomic analysis, the sequence data of isolated endophytic fungi were submitted to GenBank for Accession Number verification.
RESULTS AND DISCUSSION
Isolates and taxonomy
According to the phylogenetic results and the morphological features, 2new genera (Gibellulopsis, Plectospaerella) and 3 new species (Gibellulopsis serrate, Plectospaerella niemeijerarum, Plectospaerella oratosquillae) for the family Plectosphaerellaceae is proposed in this study, are documented in the Iraqi Mycobiota, Basrah for the first time (Table 1). The new species differs morphologically from other Plectosphaerella species by irregular chlamydospores, and the dimensions of phialides and conidia, we found chlamydospores in all of the isolates that we studied, unless Plectosphaerella oratosquillae the Chlamydospores were absent (Figure 2). The family concept was based on the holomorphic species Plectosphaerella cucumerina, which produces perithecia with elongated necks. The asexual morphs were described as phialidic with mononematous conidiophores 5. Although Plectosphaerella spp. were initially isolated from plants (from healthy or symptomatic tissue), subsequent studies found that they are also widely distributed on soils and do not necessarily exhibit host specificity 7,8,9. P. oratosquillae isolated from animals and exhibits host specificity and from soil in Germany 21,22.
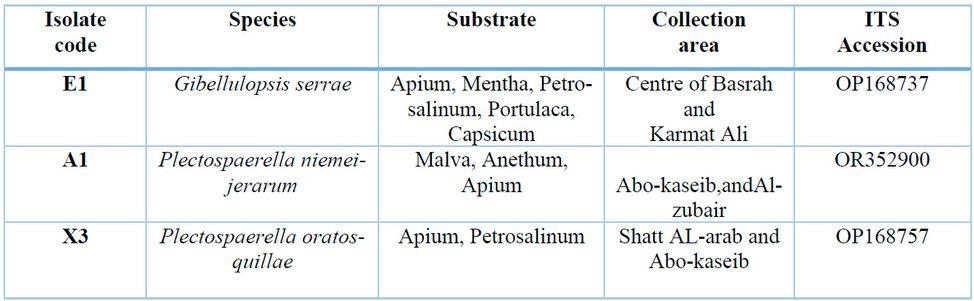
Table 1 Details isolated fungi areas, vegetable sources and ITS Accession numbers.
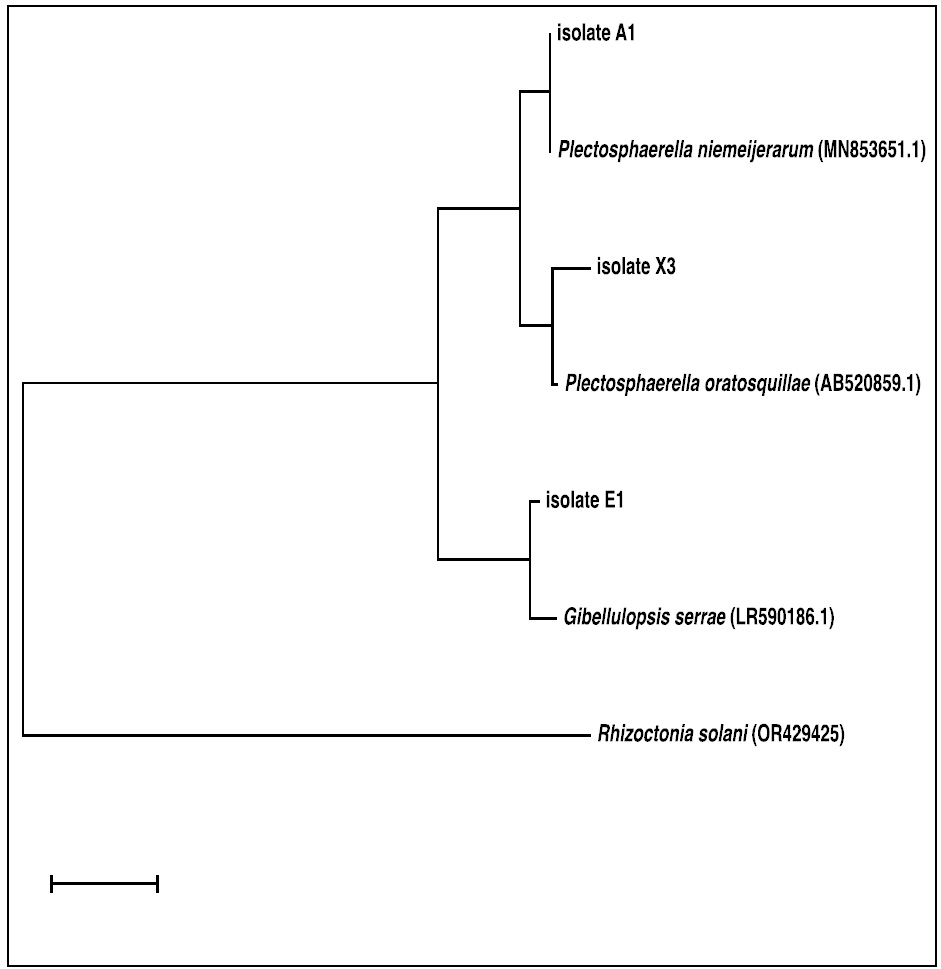
Figure 2: The phylogenetic tree represents a neighbor-joining analysis of ITS domain sequences depicting the relationships of five isolated endophytic fungi (isolate_No.X3, A1 and B1) with closely related reference sequences of Plectosphaerella species retrieved from NCBI. Each numerical value represents the percentage of bootstrap samples, a total of 1000 samples, that support the internal branches with a confidence level of 50% or higher.
Taxonomy
Plectosphaerellaceae W. Gams et al., Nova Hedwigia 85: 476. 2007. Emended. Type genus: Plectosphaerella Kleb.
Gibellulopsis Bat. and H. Maia, Anais Soc. Biol. Pernambuco 16: 153. 1959.
Gibellulopsis serrae (Maffei) Giraldo Lo pez and Crous, comb. nov. MycoBank MB828040.
Basionym: Cephalosporium serrae Maffei, Atti Ist. Bot. Pavia. Ser. 4: 196. 1930.
Culture characteristics: After seven days from incubation at 25 °C: On PDA reaching 5 - 6 cm in diameter, flat, felt or floccose, completely white color and colorless to the periphery, reverse uncolored or little yellow. On PCA, it is entirely pink in color, undiluted a puff from the medium, and the reverse is pink to brown, reaching (4 - 5) cm in diameter in seven days (Figure 3).
Description: Conidia ellipsoidal to cylindrical with rounded ends, 1-celled, hyaline, thin and smooth-walled (1 - 4 x 0.5 - 2) μm, arranged in slimy heads. Chlamydospores are mostly intercalary, singly or in pairs, globose to subglobose with a truncate base, pale green, smooth- and thick-walled (5 – 7 x 2 - 5) μm. Sexual morph not observed.
Material examined: This fungus was isolated from five samples of the plants (Apium, Mentha, Portulaca, Capsicum, Petrosalinum) collected from (the Centre of Basrah and Karmat Ali) in Basrah province. NCBI confirmed the GenBank accession number (OP168737) of this isolate. This fungus is the first record of EF fungi in Iraq. It was isolated from Dutch soil in Canada in 2007 5.
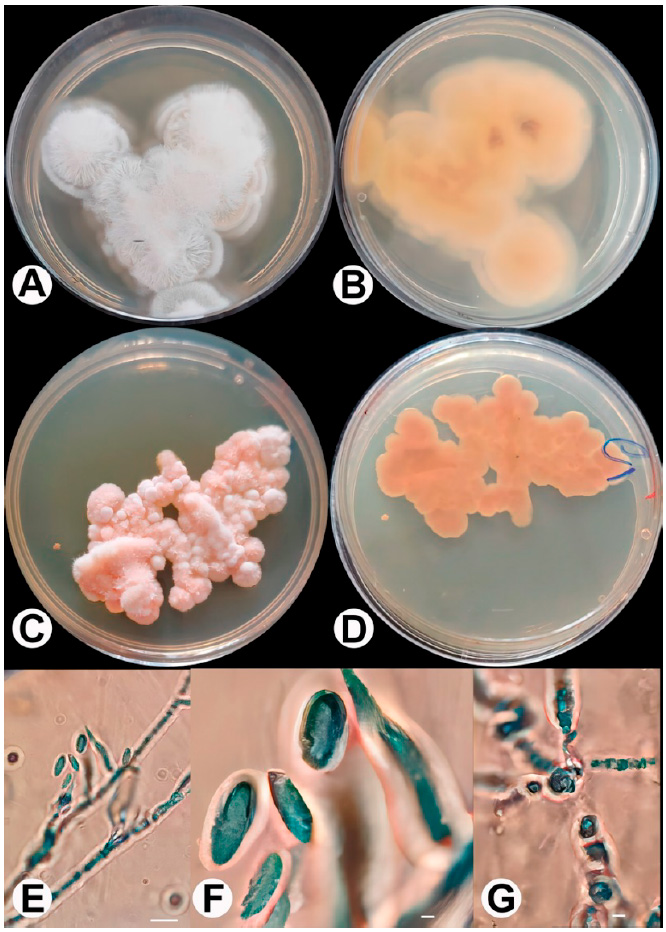
Figure 3: Gibellulopsis serrae (E1). A: G. Serra on PDA front; B: G. Serra on PDA back; C: G. Serra on PCA front; D: G. Serra on PCA back; E and F: Conidia and conidiophore; G: Chalmydospore. Bars: E = 100 μm; F,G = 7 μm.
Plectosphaerella niemeijerarum L. Lombard, 2017
Culture characteristics: Colonies on PDA, pink, mycelium appressed, slimy, minor, or no aerial mycelium, reaching a diameter of 3 cm after 7 days at 25 °C. On PCA, it is entirely light pink and slimy, flat, with little white aerial mycelium at the center (Figure 4).
Description: Mycelium hyaline, branched, septate, forming hyphal coils on PDA with phialides produced on the coils. Conidiophores are solitary, unbranched, hyaline, smooth, and thin-walled. Septum, phialide apex straight, sometimes crooked, gradually tapering to the apex, conidia aggregating in slimy heads, ellipsoid, hyaline, smooth, thin-walled (0-1) septate, (3-10) × (1-3) μm, . Chlamydospores are mostly intercalary, single, globose, green, smooth- and thick-walled (4-5× 2-5) μm. Sexual morph not observed.
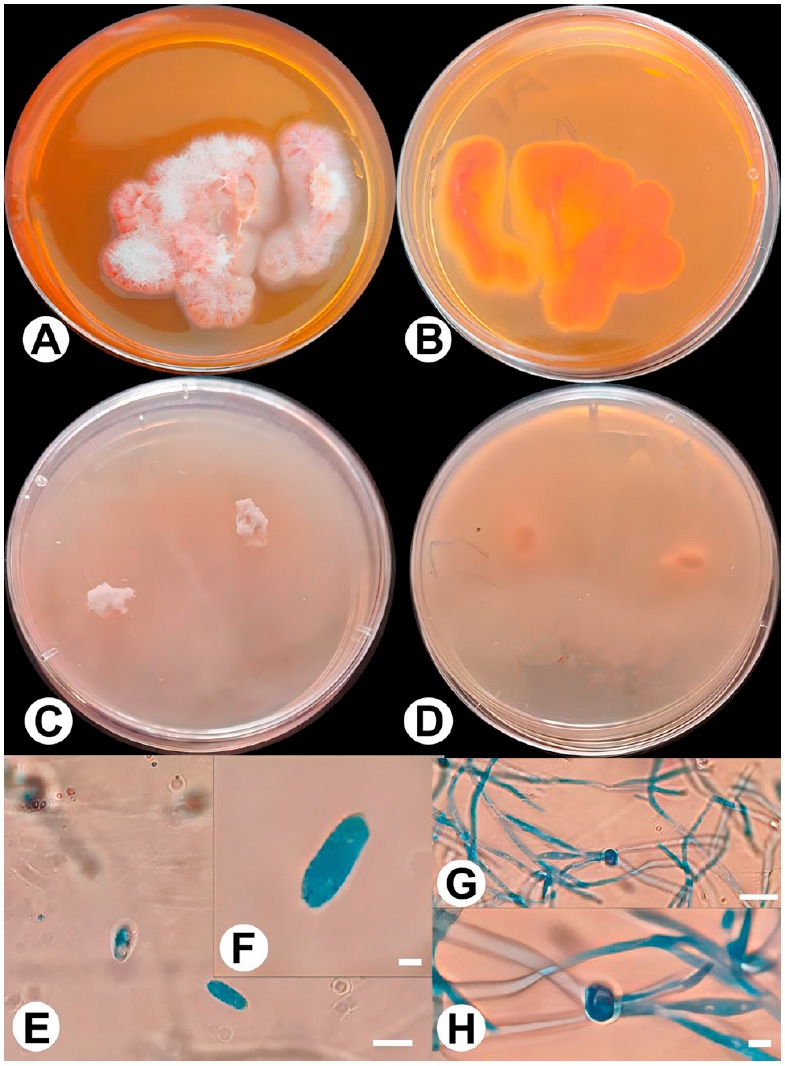
Figure 4: Plectosphaerella niemeijerarum (A1). A: P. niemeijerarum on PDA front; B: P. niemeijerarum on PDA back; C: P. niemeijerarum on PCA front; D: P. niemeijerarum on PCA back; E and F: Conida; G and H: Chalmedospore. Bars: E and G = 110 μm; F and H = 6 μm.
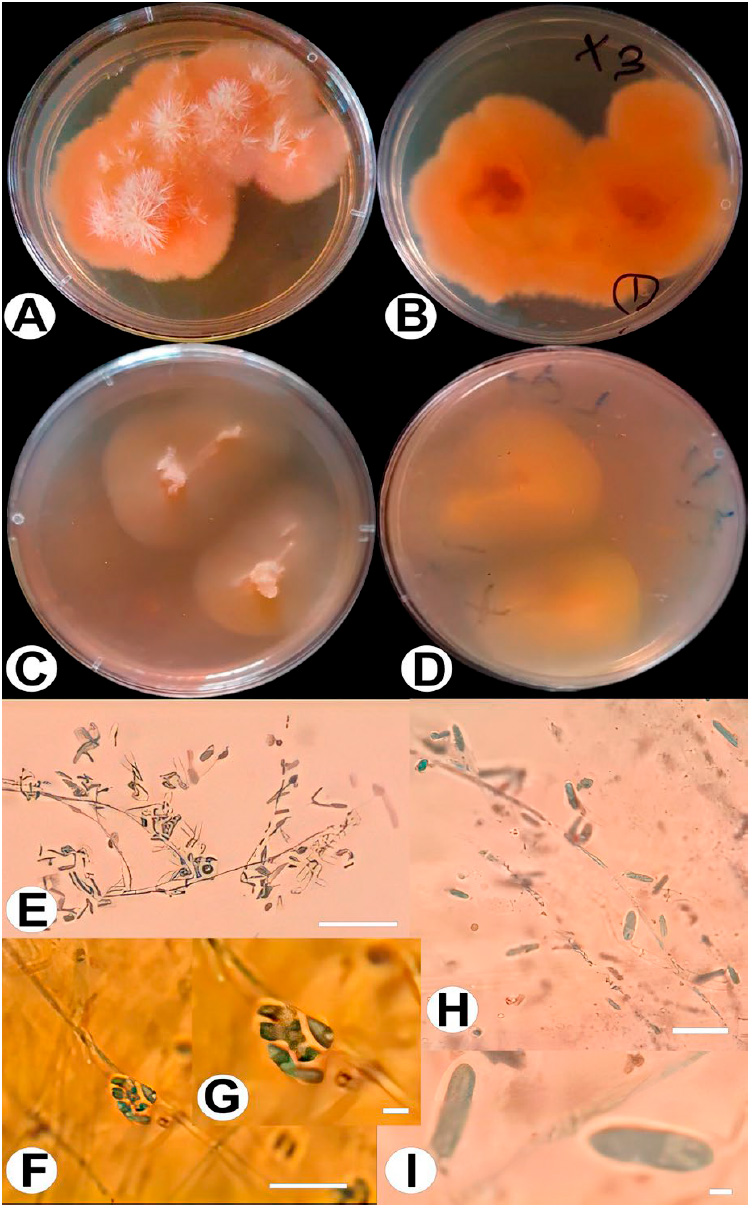
Figure 5: Plectosphaerella oratosquillae (X3). A: P. oratosquillae on PDA front; B: P. oratosquillae on PDA back; C: P. oratosquillae on PCA front; D: P. oratosquillae on PCA back; E and I: conidiaand conidiophore; F and G: aggregate in slimy head. Bars: E = 90 μm; F = 110 μm; G = 6 μm.
Material examined: This fungus was isolated from three samples of the plants (Anethum, Apium, Malva), which were collected from two areas (Abo-case, Al-Zubair) in Basrah Province. NCBI confirmed the GenBank accession number (OR429428) of this isolate. This fungus is the first record of EF fungi in Iraq and the first record on this substrate. In Italy, another kind of Plectosphaerella as a plant pathogen on basil and parsley is Plectosphaerella cucumerina, P. pauciseptata, P. plurivora, and P. ramiseptata 8. In Iran, isolated P. cucumerina as a plant pathogen causes melon decline 23. Recently, P. niemeijerarum was isolated from the Korean medical plant Ligusticum chuanxiong and another fungus, Pithomyces chartarum 24.
Plectosphaerella oratosquillae (P.M. Duc, Yaguchi and Udagawa) A.J.L. Phillips, A. Carlucci and M.L. Raimondo, comb. nov. — MycoBank MB564577
Basionym. Plectosporium oratosquillae P.M. Duc, Yaguchi and Udagawa, Mycopathologia 167: 237. 2009.
Culture characteristics: After seven days from incubation at 25 °C On PDA, Colonies were buff and salmon pink, and the mycelium was slimy with white aerial mycelium at the center, reaching 6 cm in diameter. Reverse pink color; after 14 days at 25 °C, the colonies become white and slimy. White aerial mycelium at the center, reaching 8 cm in diameter, on PCA, completely white to light pink and slimy to flat, with white aerial mycelium at the center, the reverse color (Figure 5).
Description: The conidiophores were found to be solitary, unbranched or rarely irregularly branched, and the Conidiogenous cells were monophialidic, hyaline, solitary, straight or crooked. Occasionally, the conidiogenous cells showed one septum near the base. Conidia hyaline, elliptical to ovoid, 1-2 septate (2-10 x 1-2). they aggregated in slimy heads. Chlamydospores were absent.
Material examined: This fungus was isolated from two samples of the plants (Apium, Petrosalinum), which were collected from two areas (Abo-Kaseib and Shatt-AlArab) in Basrah Province. NCBI confirmed the GenBank accession number (OP168741) of this isolate. This fungus is the first record of EF fungi in Iraq and the first record on this substrate. Most species of Plectosphaerella isolated from soil or plants except P. oratosquillae isolated from mantis shrimp (Oratosquilla oratorio) in Yamaguchi and Aichi Prefectures, Japan, are described as the new species Plectosporium oratosquillae. Thus, fungi can only be isolated from animals and exhibit host specificity 11.
CONCLUSIONS
In conclusion, the present study marked the initial investigation of the family Plectosphaerellaceae as endophytic fungi in Basrah, Iraq, particularly vegetable samples. The isolated species exhibited a wide range of morphological features, indicating that more than relying on morphological characteristics for identification may be needed and should be supplemented with modern phylogenetic techniques. These three identified species, namely Gibellulopsis serrae, Plectosphaerella niemeijerarum, and Plectosphaerella oratosquillae, were documented for the first time in the Iraqi mycobiota.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
We thank the University of Basrah, College of Science, and Department of Biology for their support and Dr Marwan Al-Maqtoofi's distinctive assistance.
REFERENCES
1. Wen J, Okyere SK, Wang S, Jianchen W, Lei X, Yinan R, Yanchun H (2022). Endophytic Fungi: An Effective Alternative Source of Plant-Derived Bioactive Compounds for Pharmacological Studies. J Fungi (Basel) 8(2):205. doi:10.3390/jof8020205
2. Rajamanikyam M, Vadlapudi V, Amanchy R, Upadhyayula SM. Endophytic Fungi as Novel Resources of Natural Therapeutics. Braz arch biol technol. 2017;60:e17160542. doi:10.1590/1678-4324-2017160542
3. Selim K, El-Beih A, Abdel-Rahman T, El Diwany A. Biology of Endophytic Fungi. CREAM. 2012;2:31-82. doi:10.5943/cream/2/1/3
4. Petrini O. Fungal Endophytes of Tree Leaves. In: Andrews JH, Hirano SS, eds. Microbial Ecology of Leaves. Brock/Springer Series in Contemporary Bioscience. New York, NY: Springer; 1991:179-197. doi:10.1007/978-1-4612-3168-4_9
5. Zare RG. Gibellulopsis is a suitable genus for Verticillium nigrescens, and Musicillium is a new genus for V. theobromae. Nova Hedwigia. 2007:463-489. doi:10.1127/0029-5035/2007/0085-0463
6. Yang XQ, Ma SY, Peng ZX, Wang ZQ, Qiao M, Yu Z. Diversity of Plectosphaerella within aquatic plants from southwest China, with P. endophytica and P. sichuanensis spp. nov. MycoKeys. 2021;80:57-75. doi:10.3897/mycokeys.80.64624
7. Giraldo A, Crous PW. Inside Plectosphaerellaceae. Stud Mycol. 2019;92:227-286. doi:10.1016/j.simyco.2018.10.005
8. Raimondo ML, Carlucci A. Characterization and pathogenicity of Plectosphaerella spp. collected from basil and parsley in Italy. Phytopathologia Mediterranea. 2018;57(2):284-295.
9. Carlucci A, Raimondo ML, Santos J, Phillips AJL. Plectosphaerella species associated with root and collar rots of horticultural crops in southern Italy. Persoonia. 2012;28:34-48. doi:10.3767/003158512X638251
10. Su L, Deng H, Niu YC. Phylogenetic analysis of Plectosphaerella species based on multi-locus DNA sequences and description of P. sinensis sp. nov. Mycological Progress. 2017;8(16):823-829. doi:10.1007/s11557-017-1319-8
11. Duc PM, Hatai K, Kurata O, et al. Fungal infection of mantis shrimp (Oratosquilla oratoria) caused by two anamorphic fungi found in Japan. Mycopathologia. 2009;167(5):229-247. doi:10.1007/s11046-008-9174-4
12. Al-Rifaie AA, Ameen MKM. New endophytic fungal species of Chaetomiaceae (Ascomycota) in Iraq. Biodiversi. 2023;24(10): 5270-5277.DOI: 10.13057/biodiv/d241007
13. Al-Dossary MA, Raheem SS, Almyah MK. 2021. Molecular identification of five species of family Chaetomiaceae (Sordariomycetes, Ascomycota) from Iraqi soil. Biodiversi. 22 (3): 1277-1284. DOI: 10.13057/biodiv/d220325.
14. Al-Rifaie AA, Al-Maqtoofi MY. Immunodetection and risk assessment for Aspergillus contamination in nuts using a highly specific monoclonal antibody. Biomedical Research. 2018;29(21). doi:10.4066/biomedicalresearch.29-18-1098
15. Mohammed-Ameen MK, Minati MH, Abbas M. Morphogenetic identification, description and pathogenicity of novel pathogens on Iraqi wheat plant (Triticum aestivum) causing head blight and crown rot diseases. Biodiversi. 2021;22(5). doi:10.13057/biodiv/d220565
16. Minati MH, Mohammed-Ameen MK. Novel report on six Fusarium species associated with head blight and crown rot of wheat in Basra province, Iraq. Bull Natl Res Cent. 2019;43(1):139. doi:10.1186/s42269-019-0173-z
17. Ibrahim M, Oyebanji E, Fowora M, et al. Extracts of endophytic fungi from leaves of selected Nigerian ethnomedicinal plants exhibited antioxidant activity. BMC Complementary Medicine and Therapies. 2021;21(1):98. doi:10.1186/s12906-021-03269-3
18. White TJ, Bruns T, Lee S, Taylor JW, others. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: A Guide to Methods and Applications. Vol 18. Academic Press, Inc.; 1990:315-322.
19. Al-Maqtoofi M, Thornton CR. Detection of human pathogenic Fusarium species in hospital and communal sink biofilms by using a highly specific monoclonal antibody. Environ Microbiol. 2016;18(11):3620-3634. doi:10.1111/1462-2920.13233
20. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725-2729. doi:10.1093/molbev/mst197
21. Muñoz-Barrios A, Sopeña-Torres S, Ramos B, et al. Differential Expression of Fungal Genes Determines the Lifestyle of Plectosphaerella Strains During Arabidopsis thaliana Colonization. MPMI. 2020;33(11):1299-1314. doi:10.1094/MPMI-03-20-0057-R
22. Wahdan SF, Buscot F, Purahong W. Future Climate Alters Pathogens-Microbiome Co-occurrence Networks in Wheat Straw Residues during Decomposition. Proceed. 2021;66(1):22. doi:10.3390/proceedings2020066022
23. Mirtalebi M, Sabahi F, Banihashemi Z. First report of melon decline caused by Plectosphaerella cucumerina in Iran. Mycolog. Iran. 2022;9(1): 97–103. DOI: 10.22043/MI.2022.357797.1210
24. Park H, Jung CR, Eom AH. Two Unrecorded Enophytic Fungi Isolated from Root of Ligusticum chuanxiong in Korea: Pithomyces chartarum and Plectosphaerella niemeijerarum. The Kor. J of Myco. 2019;47(4):329-334. doi:10.4489/KJM.20190038
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 February 2024
Citation: Al-Rifaie A. A. and Mohammed-Ameen M.K. Morphogenetic identification of new endophytic fungal species related to the family of Plectosphaerellaceae in Iraq. Bionatura Journal 2024; 1 (1) 36. http://dx.doi.org/10.21931/BJ/2024.01.01.36
Additional information Correspondence should be addressed to [email protected]
Peer review information. Bionatura Journal thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura Journal stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
