2024..01.01.72
Files > Conference Series > 2024 > Chimboazo ild pagina nueva
Isolation, characterization and identification of klebsiella sp. 35-DOR_27F isolated from textile effluents
Derly Ortiz-Romero1, Daniela Camacho-Valencia2,3, Stamber Alvaro Ramírez-Revilla2,3*
1Universidad Católica de Santa María, Arequipa, Perú; [email protected].
2Universidad Tecnológica del Perú, Arequipa, Perú; [email protected].
3Laboratorio de Calidad de Agua y Medio Ambiente, Universidad Tecnológica del Perú; [email protected].
* Correspondence: [email protected]; Tel.: +51974434314
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.72
ABSTRACT
This research study aimed to isolate and characterize a new bacterial strain from textile effluents. In order to do this, bacteria were cultured using the MSM medium, where a colony was isolated through six successive pickings. It then underwent a DNA extraction process using the phenol-chloroform-isoamyl alcohol methodology, and an electrophoresis was carried out to confirm the extraction. In addition, the isolated colony was identified
Keywords: Bacterial strain, Klebsiella sp., DNA extraction, decolorization, effluents.
INTRODUCTION
One of the main problems affecting the world's environment is water contamination due to industrial activity, which generates high amounts of polluting effluents that produce toxic effects. 1. For example, the textile industry generates wastewater containing artificial recalcitrant or carcinogenic dyes 2-4.
Worldwide, around 10,000 types of dyes and pigments are used in a wide range of diverse industries, and approximately 7 × 105 tons of them are produced annually due to the industrialization and demand of the textile industry 5,6. Among the dyes used in the industry, around 20 % remain unused (they do not fully penetrate the fabrics) and are released directly into the sewage system. This renders the water unfit for use and impedes light penetration into bodies of surface water, adversely affecting the aquatic environment 7-9.
Industrial dyes contain various structures, most composed of three reactive groups: azo, anthraquinon and phthalocyanin 6,10,11. Numerous research studies have been carried out to perfect a treatment that allows the degradation of these textile dyes and colorings and the recovery of the natural water resource. A viable alternative is biological treatment since it is inexpensive, ecological, and functional compared to other chemical or physical treatments that require much energy 6,11,12.
Some studies have used different bacteria for the biodegradation of reactive textile dyes, such as: Alcaligenes faecalis, Bacillus cereus and Bacillus sp. 6; Lysinibacillus sphaericus and Aeromonas hydrophila 11; Micrococcus luteus and Bacillus subtilis 4; Staphylococcus sp. 13; Citrobacter freundii 1; Planococcus and Bacillus 14, Pseudomonas 15,16; Aeromonas 17, Bacillus and Lysinibacillus 18; Paenibacillus polymyxa, Micrococcus luteus and Micrococcus 19. Other studies have used microorganisms such as Galactomyces geotrichum 20, Phanerochaete chrysosporium 21 and Enterobacter 22 for the same purpose.
This study focused on isolating, characterizing, and identifying a new bacterial strain from industrial textile dyes effluents. The strain was genetically assessed by 16S rRNA analysis and software. Finally, the isolated bacteria were grown in a culture medium enriched with an azoic dye, and their decolorization capacity was evaluated.
MATERIALS AND METHODS
Culture medium
The mineral salts medium (MSM) consisted of the following components in (g/L):
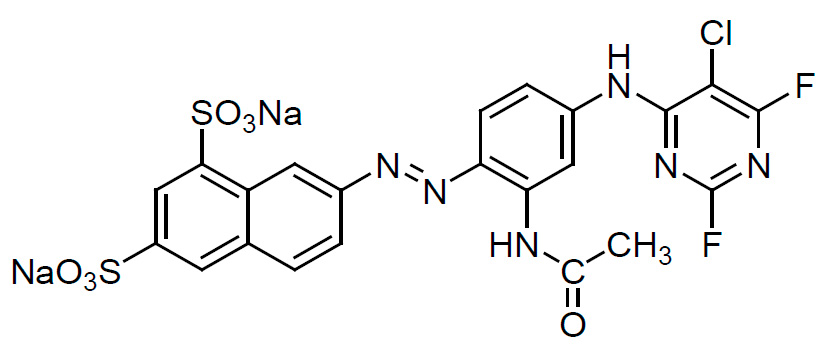
Figure 1: Chemical structure of the Golden Yellow K2R dye.
Obtaining the sample
A sample of textile effluent was taken to the laboratory to isolate the bacteria. 1.5 mL of effluent was placed in each of the 10 Eppendorf tubes before being taken to the microcentrifuge and run at 10,000 RPM for 5 min. The supernatant was eliminated, and the pellets formed in each tube were gathered. Subsequently, 100 µL of the sample was seeded using a Drigalsky loop on a plate with MSM agar and the Golden Yellow K2R dye as the only carbon source. The sample was incubated at 37°C for 24 hours.
Picking
A colony was removed from the initial seeding. It was placed in an Eppendorf tube with 1 mL of Triton and vortexed for 5 min. Then, it was seeded with a cole loop on a new plate. A total of 6 pickings were made following the same procedure.
Antibiogram
To discover the sensitivity of the isolated bacteria to different antibiotics, a sample (of the pure culture in MSM broth) was seeded on nutritive agar using a sterile swab. Then, paper discs with the chosen antibiotics for
DNA extraction
An Eppendorf tube was filled with 100 µL of microbeads and 500 µL of sterile water, and a loop of bacteria was dissolved. Then, it was vortexed at maximum speed for 5 min. After that, 500 µL of a phenol-chloroform-isoamyl alcohol mixture was added during the vortexing, and lastly, it was placed in the microcentrifuge at 15,000 rpm for 10 min. The supernatant was recovered by adding 600 µL of isopropyl alcohol, mixed gently and then centrifuged at 15,000 rpm for 10 min. After discarding the supernatant, ethanol was applied to dry the pellet, and finally, it was re-suspended with 50 µL of sterile water, keeping the DNA at 4 °C. The technique used is a variant of the optimal technique for bacterial DNA isolation by bead beating described by Fujimoto et al. 25.
Sequencing the 16S rRNA gene of the 35-DOR_27F bacteria
Once the DNA extraction had been verified, the 16S rRNA gene was amplified by the PCR method. The obtained product was purified and sent for sequencing to the Instituto de Biotecnología de ADN de Uchumayo using the FinchTV software. Sequencing analysis of the 16S rRNA gene was performed at the Laboratory of Molecular Biology, Harvard University, Cambridge, MA, USA.
Bioinformatic analysis by NCBI BLAST
For the bioinformatic analysis, the NCBI BLAST database was used. The previously obtained DNA sequence was inserted. Likewise, in order to obtain the phylogenetic tree, the BioEdit, Mega and Trie View Explorer software were used. The 16S rRNA gene sequence of bacterial strain 35-DOR_27F was inserted into the NCBI GenBank database for alignment and comparison with stored bacterial sequences to assess their degree of homology using the BLASTn algorithm 26. BioEdit, Mega and Trie View Explorer software were used to obtain the phylogenetic tree.
Biotechnological potential studies
100 µL of the isolated bacterial strain was inoculated in 10 mL MSM broth with Golden Yellow K2R dye (50 mg/L). Incubation was carried out at 37 °C for 72 hours. Afterward, the broth was centrifuged at 6000 rpm for 20 min. Then, the supernatant was separated, and its absorbance was measured at 388 nm (corresponding to the λmax of the dye) in a Thermogenesis UV–visible spectrophotometer. For the biodegradation study, an Agilent Technologies Cary 60 UV-V spectrophotometer was used to quantify the absorbance of a 60 mg/L solution of the yellow dye K2R. The isolated strain was cultured in peptonized broth for 48 h at 37°C, and then 1 mL of inoculum was taken and placed in a flask containing 100 mL of a 50 mg/L solution of the dye in peptonized water. The percentage of decolorization was evaluated after 72 hours using Equation 1:
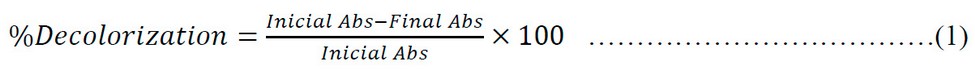
RESULTS
Morphological and cultural characteristics
Gram-negative bacteria were isolated with the following characteristics: circular point shape, whole border, flat colony elevation, creamy consistency, smooth surface and transparent white pigmentation.
Antibiogram
Results of the susceptibility of the isolated bacteria are shown in Table 1. After 24 hours of incubation, the bacteria were sensitive to most chosen antibiotics except ampicillin/sulbactam. Amikacin was the antibiotic that presented the largest halo, indicating that the isolated bacteria have a greater sensitivity to said antibiotic.
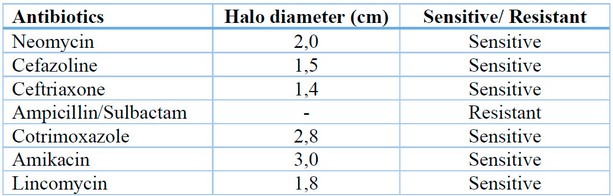
Table 1. Samples resistance to antibiotics.
Figure 2 shows the inhibition halos for the different antibiotics with the isolated bacteria on nutrient agar. The biggest inhibition halo corresponds to the antibiotic amikacin.
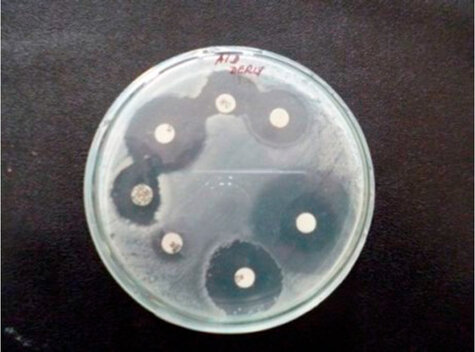
Figure 2: Antibiogram of the isolated bacteria.
The sequence of the 16S rRNA gene of the 35-DOR_27F bacteria
Using the FinchTV software, it was possible to observe the sequence of the public and pyrimidine bases of the extracted DNA, which is shown in Figure 3.
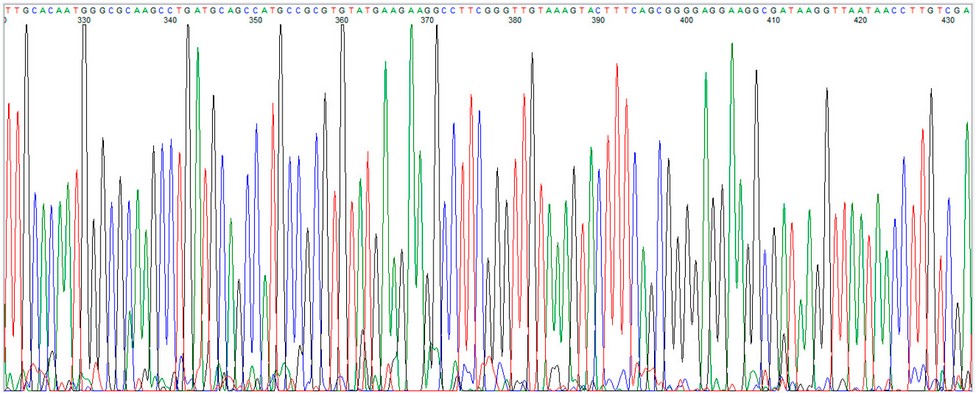
Figure 3: Visualization of the bacterial DNA sequence using FinchTV software.
Bioinformatic analysis using NCBI BLAST
After the sequence was obtained and the bioinformatic analysis using BLAST was performed, it was deter-mined that the isolated strain shares a high percentage of similarity (99 %) with Klebsiella sp., which is cata-loged as a species capable of decolorizing textile effluents. Figure 4 shows the results of the BLAST analysis with the greatest similarity to Klebsiella sp., and Figure 5 shows the phylogenetic tree; it is notable that the bacterium encoded as 35-DOR_27F is found at the same height as Klebsiella sp., which indicates it could belong to this species.
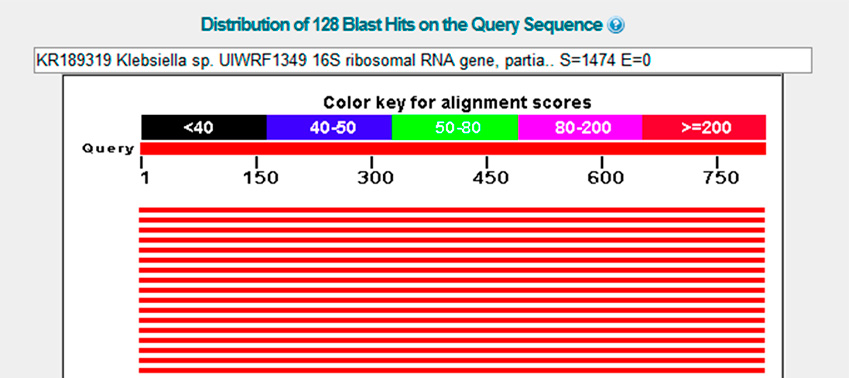
Figure 4: Analysis of the isolated bacteria using BLAST.
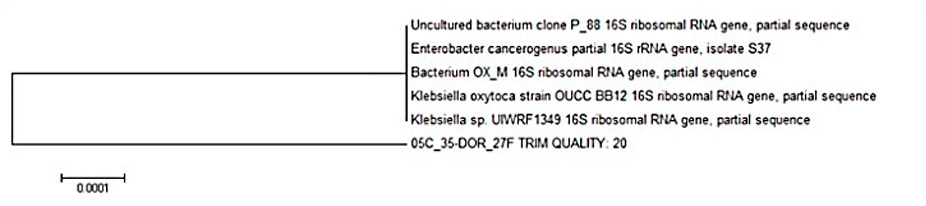
Figure 5: Phylogenetic tree of the isolated bacterium.
Biotechnological potential studies
The wavelength of maximum absorption for a 60 mg/L solution of Golden Yellow K2R was 388 nm with an absorbance of 0.9522. Figure 6 presents the spectrum obtained.
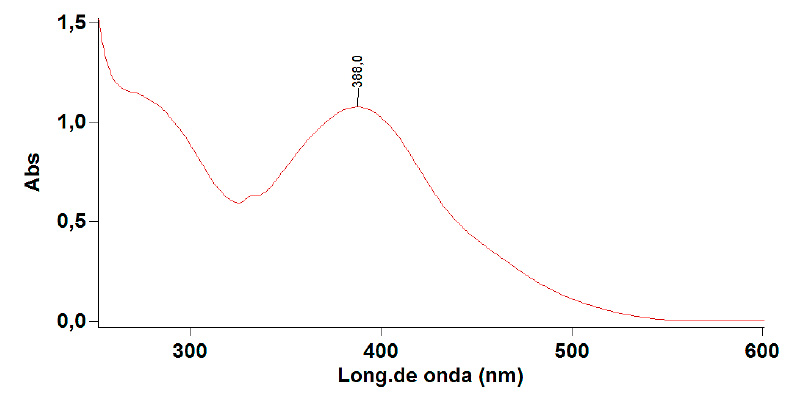
Figure 6: Spectral scanning of the Gold Yellow K2R.
After 72 hours of incubation, the results indicated that the decolorization of Golden Yellow K2R by the isolated strain identified as Klebsiella sp. was 74 %, suggesting that the isolated bacteria could be used in bioremediation processes.
DISCUSSION
Different bacterial species have been isolated from textile sources, demonstrating their bioremediation capac-ity against azoic dyes in solution. Bacillus and Alcaligenes stand out among the identified genera, as they are efficient when treating Novacron Super Black G dye 6. In addition, Alcaligenes aquatilis proved effective in degrading Synazol Red 6HBN colorant (82 %), suggesting it may be helpful to in wastewater treatment 27. Furthermore, Bacillus subtilis, Bacillus megaterium, Erysipelothrix and Amphibacillus xylanus were isolated and identified, and their degradation ability was studied against 7 azoic dyes. Results demonstrated that the dyes were decolorized (at least 50 %) after 3 days of incubation. However, none of the isolates could decol-orize Novacron Red 28. Moreover, Alishewanella sp. has also shown decolorization capacity against Sumifex Tourqi blue (83 %) after 6 days, thus showing its potential for removing azoic dyes 29. In the present research study, the isolated bacterial strain corresponds to Klebsiella sp., which was grown on MSM broth enriched with Golden Yellow K2R azoic dye and showed a decolorization of 74 %. Other studies report bacterial growth in agar enriched with different azoic dyes, such as Bezema Yellow S8-G and Bezema Red S2-B, showing decolorization rates of up to 90 % 8.
CONCLUSIONS
A new bacterium obtained from textile effluents was isolated, and the code 35-DOR_27F was assigned. Sequencing the 16S rRNA gene was found to correspond to Klebsiella genus. Furthermore, after 72 hours of incubation, it exhibited a decolorization percentage of 74 % of the Golden Yellow K2R dye. This
the study indicates that this bacterium has promising characteristics for addressing pollution caused by textile effluents.
Author Contributions: "Conceptualization, D. Ortiz-Romero; methodology, D. Ortiz-Romero and D. Camacho-Valencia; software, D. Ortiz-Romero.; validation, S.A Ramírez- Revilla; formal analysis, D. Ortiz-Romero and D. Camacho-Valencia; investigation, D. Ortiz-Romero and S.A Ramírez- Revilla; data curation, D. Ortiz-Romero and S.A Ramírez- Revilla; writing—original draft preparation, D. Ortiz-Romero and D. Camacho- Valencia; writing—review and editing, D. Ortiz-Romero and S.A Ramírez- Revilla; supervision, S.A Ramírez- Revilla; project administration, D. Ortiz-Romero and S.A Ramírez- Revilla.; funding acquisition, S.A Ramírez- Revilla. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
1. Sunkar S, C. VN, K. R. Citrobacter freundii mediated degradation of textile dye Mordant Black 17. Journal of Water Process Engineering. 2015;8:28-34. doi:10.1016/j.jwpe.2015.08.006
2. Chung KT. The Significance of Azo-Reduction in the Mutagenesis and Carcinogenesis of Azo Dyes. Vol 114.; 1983.
3. Kawakami T, Isama K, Nakashima H, Tsuchiya T, Matsuoka A. Analysis of primary aromatic amines originated from azo dyes in commercial textile products in Japan. J Environ Sci Health A Tox Hazard Subst Environ Eng. 2010;45(10):1281-1295. doi:10.1080/10934529.2010.493827
4. Ito T, Shimada Y, Suto T. Potential use of bacteria collected from human hands for textile dye decolorization. Water Resour Ind. 2018;20:46-53. doi:10.1016/j.wri.2018.09.001
5. Aksu Z, Tezer S. Biosorption of reactive dyes on the green alga Chlorella vulgaris. Process Biochemistry. 2005;40(3-4):1347-1361. doi:10.1016/j.procbio.2004.06.007
6. Hossen MZ, Hussain ME, Hakim A, Islam K, Uddin MN, Azad AK. Biodegradation of reactive textile dye Novacron Super Black G by free cells of newly isolated Alcaligenes faecalis AZ26 and Bacillus spp obtained from textile effluents. Heliyon. 2019;5(7). doi:10.1016/j.heliyon.2019.e02068
7. Valli Nachiyar C, Namasivayam SKR, Kumar RR, Sowjanya M. Bioremediation of textile effluent containing Mordant Black 17 by bacterial consortium CN-1. Journal of Water Process Engineering. 2014;4(C):196-200. doi:10.1016/j.jwpe.2014.10.003
8. Karim ME, Dhar K, Hossain MT. Decolorization of Textile Reactive Dyes by Bacterial Monoculture and Consortium Screened from Textile Dyeing Effluent. Journal of Genetic Engineering and Biotechnology. 2018;16(2):375-380. doi:10.1016/j.jgeb.2018.02.005
9. Garzón-Pérez AS, Paredes-Carrera SP, Martínez-Gutiérrez H, Cayetano-Castro N, Sánchez-Ochoa JC, Pérez-Gutiérrez RM. Effect of combined microwave-ultrasound irradiation in the structure and morphology of hydrotalcite-like compounds al/mg-ch3coo and its evaluation in the sorption of a reactive dye. Revista Mexicana de Ingeniera Quimica. 2020;19(1):363-375. doi:10.24275/rmiq/Mat567
10. Axelsson J, Nilsson U, Terrazas E, Alvarez Aliaga T, Welander U. Decolorize the Reactive Red 2 and Reactive Blue 4 textile dyes using Bjerkandera sp. Strain BOL 13 in a continuous rotating biological contactor reactor. Enzyme Microb Technol. 2006;39(1):32-37. doi:10.1016/j.enzmictec.2005.09.006
11. Kumar SS, Shantkriti S, Muruganandham T, Murugesh E, Rane N, Govindwar SP. Bioinformatics aided microbial approach for bioremediation of wastewater containing textile dyes. Ecol Inform. 2016;31:112-121. doi:10.1016/j.ecoinf.2015.12.001
12. Saratale RG, Saratale GD, Chang JS, Govindwar SP. Bacterial decolorization and degradation of azo dyes: A review. J Taiwan Inst Chem Eng. 2011;42(1):138-157. doi:10.1016/j.jtice.2010.06.006
13. Durai S, Kumar NM, Saravanan D. Isolation of dye degrading bacteria from textile effluent. Available online www.jocpr.com Journal of Chemical and Pharmaceutical Research. 2015;7(3):2214-2218. https://www.researchgate.net/publication/305787431
14. Shah MP. Isolation and Screening of Dye Decolorizing Bacteria. J Appl Environ Microbiol. 2014;2(5):244-248. doi:10.12691/jam-2-5-7
15. Jadhav JP, Kalyani DC, Telke AA, Phugare SS, Govindwar SP. Evaluation of the efficacy of a bacterial consortium for the removal of color, reduction of heavy metals, and toxicity from textile dye effluent. Bioresour Technol. 2010;101(1):165-173. doi:10.1016/j.biortech.2009.08.027
16. Hussain S, Maqbool Z, Ali S, et al. Biodecolorization of reactive black-5 by a metal and salt tolerant bacterial strain Pseudomonas sp. RA20 isolated from Paharang drain effluents in Pakistan. Ecotoxicol Environ Saf. 2013;98:331-338. doi:10.1016/j.ecoenv.2013.09.018
17. Du LN, Li G, Zhao YH, et al. Efficient metabolism of the azo dye methyl orange by Aeromonas sp. strain DH-6: Characteristics and partial mechanism. Int Biodeterior Biodegradation. 2015;105:66-72. doi:10.1016/j.ibiod.2015.08.019
18. Anjaneya O, Souche SY, Santoshkumar M, Karegoudar TB. Decolorization of sulfonated azo dye Metanil Yellow by newly isolated bacterial strains: Bacillus sp. strain AK1 and Lysinibacillus sp. strain AK2. J Hazard Mater. 2011;190(1-3):351-358. doi:10.1016/j.jhazmat.2011.03.044
19. Moosvi S, Kher X, Madamwar D. Isolation, characterization and decolorization of textile dyes by a mixed bacterial consortium JW-2. Dyes and Pigments. 2007;74(3):723-729. doi:10.1016/j.dyepig.2006.05.005
20. Govindwar SP, Kurade MB, Tamboli DP, Kabra AN, Kim PJ, Waghmode TR. Decolorization and degradation of xenobiotic azo dye Reactive Yellow-84A and textile effluent by Galactomyces geotrichum. Chemosphere. 2014;109:234-238. doi:10.1016/j.chemosphere.2014.02.009
21. Enayatizamir N, Tabandeh F, Rodríguez-Couto S, Yakhchali B, Alikhani HA, Mohammadi L. Biodegradation pathway and detoxification of the diazo dye Reactive Black 5 by Phanerochaete chrysosporium. Bioresour Technol. 2011;102(22):10359-10362. doi:10.1016/j.biortech.2011.08.130
22. Chen G, Huang M hong, Chen L, Chen D hui. A batch decolorization and kinetic study of Reactive Black 5 by a bacterial strain Enterobacter sp. GY-1. Int Biodeterior Biodegradation. 2011;65(6):790-796. doi:10.1016/j.ibiod.2011.05.003
23. Brilon C, Beckmann W, Hellwig M, Knackmuss3t HJ. Enrichment and Isolation of Naphthalenesulfonic Acid-Utilizing Pseudomonads.; 1981
24. Of R, Technologists. TECHNICAL SECTION ANTIBIOTIC SUSCEPTIBILITY TESTING BY A
STANDARDIZED SINGLE DISK METHOD. Vol 45.; 1966. https://academic.oup.com/ajcp/article
25. Fujimoto S, Nakagami Y, Kojima F. Optimal Bacterial DNA Isolation Method Using Bead-Beating
Technique. Vol 3.; 2004. https://www.researchgate.net/publication/268409445
26. Zhang Z, Schwartz S, Wagner L, Miller W. A Greedy Algorithm for Aligning DNA Sequences. Vol 7.
Mary Ann Liebert, Inc. Pp; 2000.
27. Ajaz M, Rehman A, Khan Z, Nisar MA, Hussain S. Degradation of azo dyes by Alcaligenes aquatilis 3c and its potential use in the wastewater treatment. AMB Express. 2019;9(1). doi:10.1186/s13568-019-0788-3
28. Mahbub KR, Mohammad A, Ahmed MM, Begum S. Decolorization of Synthetic Dyes Using Bacteria Isolated from Textile Industry Effluent. Asian Journal of Biotechnology. 2012;4(3):129-136. doi:10.3923/ajbkr.2012.129.136
29. Ajaz M, Elahi A, Rehman A. Degradation of azo dye by bacterium, Alishewanella sp. CBL-2 isolated from industrial effluent and its potential use in decontamination of wastewater. Journal of Water Reuse and Desalination. 2018;8(4):507-515. doi:10.2166/wrd.2018.065
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 February 2024
Citation: Ortiz-Romero D., Camacho-Valencia D., Ramírez-Revilla S. A. Isolation, characterization and identification of klebsiella sp. dor_27f isolated from textile effluents. Bionatura Journal 2024; 1 (1) 72. http://dx.doi.org/10.21931/BJ/2024.01.01.72
Additional information Correspondence should be addressed to [email protected]
Peer review information. Bionatura Journal thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura Journal stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
