2024..01.01.60
Files > Conference Series > 2024 > Chimborazo
Genotypic Detection of Carbapenems Resistance Genes in Acinetobacter baumannii Isolated from Urinary Tract Infection Patients
Thualfakar Hayder Hasan1, Ameer Sadeq Yasir AL-Ethari2, Sddiq Ghani Al-Muhanna3, Israa Abdul Ameer Al-Kraety4*
1jabir ibn Hayyan Medical University, Iraq, Najaf, [email protected],
2,3,4Department of Medical Laboratory Techniques, College of Health and Medical Techniques, University of Alkafeel. Najaf, Iraq
*Correspondence: [email protected],
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.60
ABSTRACT
Acinetobacter baumannii is a Gram-negative bacterium characterized by its short, round, rod-shaped morphology. It is an opportunistic pathogen that poses a significant threat, particularly to immunocompromised patients, often those with hospital stays lasting less than 90 days. Between June 2022 and July 2023, 214 urine samples were collected from individuals suspected of having urinary tract infections (UTIs). These samples were subjected to antibiotic resistance testing, focusing on detecting specific genes related to carbapenem resistance, namely blaNDM, blaKPC, and blaVIM.The study's results revealed a notable trend in antibiotic resistance among the bacterial isolates. Ceftazidime, cefotaxime, and ceftriaxone, commonly used antibiotics for UTIs, showed a high resistance rate among the tested isolates. This resistance highlights the challenges healthcare professionals face when treating UTIs caused by Acinetobacter baumannii. On the other hand, the isolates displayed a comparatively lower resistance rate to imipenem and meropenem, two necessary carbapenem antibiotics. This lower resistance to carbapenems is encouraging as these drugs are often considered the last line of defense against multidrug-resistant bacterial infections. The presence of carbapenem resistance genes, such as blaNDM, blaKPC, and blaVIM, in the Acinetobacter baumannii isolates is of particular concern. These genes confer resistance to carbapenem antibiotics, crucial for treating severe infections caused by multidrug-resistant bacteria. In conclusion, the study aims to study the growth of antibiotic resistance in Acinetobacter baumannii, especially in urinary tract infections in immunocompromised patients with more extended hospital stays. It also highlights the need for Surveys and periodic examinations to detect the spread of bacteria and their resistance.
Keywords: Carbapenems, UTI, genes, blaNDM, blaKPC, and blaVIM.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative bacterium that is small, spherical, and rod-shaped. An opportunistic bacterium 1. Regularly, it has been shown to colonize the skin, oropharynx, and respiratory tract. Recently, it has been shown that A. baumannii determines a "red alert" in hospital settings due to the emergence of antibiotic resistance 2. The phenomenon of multi-drug resistance (MDR) organism's phenomenon has progressively become a cause of severe nosocomial and community-acquired infections 3. Over time, antibiotics became less effective due to the emergence of many resistance mechanisms, such as secretion of beta-lactamase enzymes, receptor modulation, and others 4. Carbapenem antibiotics are considered one of the most effective antibiotics today and the least affected by bacterial resistance; the resistant-isotype testing showed that the isolates were effective against a wide range of bacterial illnesses 5. Carbapenems are the most potent beta-lactam antibiotics, exhibiting broad-spectrum antibacterial action against Gram-positive and Gram-negative bacteria. Combining a carbapenem and beta-lactam rings gives this compound its peculiar molecular structure6. Most beta-lactamases are immune to this mix (ESBLs) of antibiotics, including ampicillin, carbenicillin, and the extended-spectrum beta-lactamases 7. Gram-negative bacteria resistance to carbapenem is a global public health issue. Such resistance, especially when mediated by genes expressing carbapenemase, rapidly develops and generates significant outbreaks, significantly reducing the treatment options available 8. Resistance to β-lactams, notable carbapenems, is mainly caused by enzymatic breakdown by -lactamases in the Enterobacteriaceae. The NDM-1, KPC, was first identified in 2008, although NDM-1 was found in water samples from New Delhi since 2006. 9. The majority of NDM-1 infections are from the Indian subcontinent 10. Horizontal gene transfer across bacteria promotes the transmission of antibiotic-resistance genes 11. These plasmids have populations with identical structures but distinct antibiotic drug-resistance cassette compositions 12. This mechanism has allowed antibiotic resistance genes to spread quickly among Enterobacteriaceae and other pathogens like Acinetobacter baumanii 13. Antibiotic resistance genes 14, such as the extended-spectrum -lactamase CTX-M-15 in Escherichia coli ST131 and KPC in Klebsiella pneumoniae sequence type (ST) 258, may clonally propagate in successfully pathogenic strains 15,16. Due to HGT and clonal growth 17, KPC and NDM-1 quickly spread after their first appearance. Comparable mobile elements will likely make KPC and NDM-1 accessible in comparable pathogen populations 18. This is because KPC and NDM-1 have similar distributions and resistance spectra (both give resistance to practically all B-lactam antimicrobials). Clinical Enterobacteriaceae isolates from Pakistan and the United States 18 tested negative for carbapenemase, NDM-1, and KPC. This research aimed to phenotypically and genetically evaluate the antibacterial efficacy of imipenem and meropenem. The study aimed to investigate antibiotic resistance in Acinetobacter baumannii isolated from urinary tract infections (UTIs). Specific objectives included assessing resistance to common UTI antibiotics, detecting carbapenem resistance genes, and evaluating resistance to carbapenem drugs. The research focused on immunocompromised patients with shorter hospital stays. The findings provide insights into the antibiotic resistance landscape of Acinetobacter baumannii, informing strategies for UTI management.
MATERIALS AND METHODS
Isolation and identification of Acinetobacter baumanii
Urine samples were randomly collected from 214 outpatients with suspected UTIs from June 2022 to July 2023. The urine inoculates onto blood agar plates and MacConkey agar. The colony-formed unit method grows a singular, refined bacterial colony. The urine specimens that contain lower than 105 CFU / ml are eliminated. All bacteria secluded are resembled according to colony morphologic and criterion microbiological experience as colony morphologic, gram lines, oxidase experience, catalase, IMVIC tests, coagulation trial, and outgrowth in Maconkey agar (OxoidTM). The Acinetobacter baumanii suspected colonies were diagnosed depending on the vitec2 system.
Antimicrobial sensitivity test
Disk diffusion method on Muller-Hinton agar medium (Oxoid, UK) was carried out to determine the susceptibility of Acinetobacter baumannii isolates against five standard antibiotic disks including Cefotaxime (30µg), Ceftriaxone (30µg), Ceftazidime (30µg), Imipenem (10 µg), and Meropenem (10µg). MacFarland microbial suspension was used as an inoculum of approximately 1×108 CFU/ml. The cultures were incubated at 37 degrees Celsius for 18h under aerobic conditions, according to Kirby-Baur19. Zones of inhibition were calculated and interpreted as recommended by the National Committee for Clinical Laboratories Standard guidelines (Clinical and Laboratory Standards Institute,2023).
PCR for detecting genes
The DNA extraction by extraction DNA kit. The primers used in this study and the PCR thermocycling conditions are described in Tables 1 and 2 for detecting three resistance genes associated with carbapenems.

Table 1: The primer sequence used in PCR for Acinetobacter baumannii.
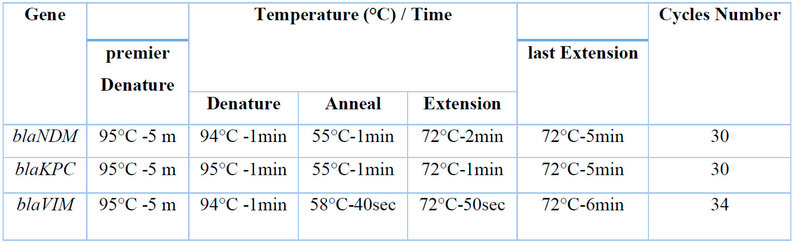
Table 2: PCR thermal cycling conditions for Acinetobacter baumannii resistance gene.
RESULTS AND DISCUSSION
Total bacterial isolates
Out of (214) collected urine samples, the result showed that 184 samples (85.98%) were G- bacterial isolates and 28 samples (13.08%) were G+ bacterial isolates. In contrast, the results showed 2 samples (0.93 %) with no growth, as shown in Figure 1.
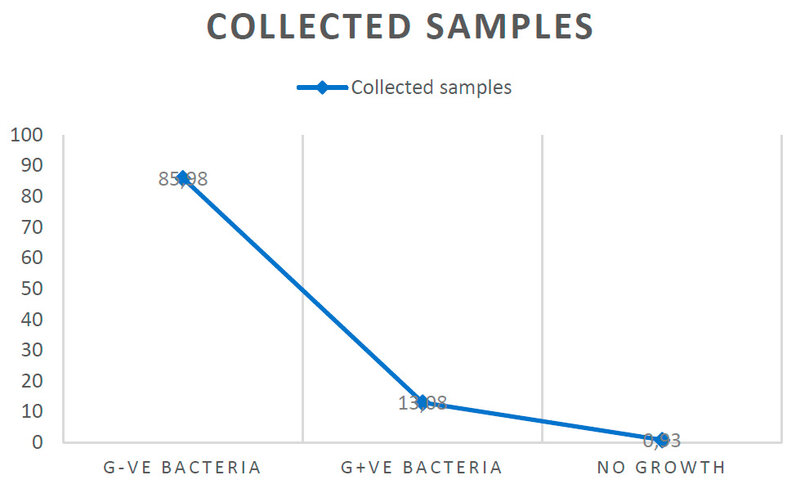
Figure 1: Percentage of total samples isolated from unhealthy with UTI.
The study results proved that the gram-negative bacterial isolates were 89 isolates (48.36%) for E. coli, 62 isolates (33.69%) for K. pneumonia, 19 isolates (10.32%) for A. bumanni, 8 isolates (0.43%) for Pseudomonas aeruginosa, and 6 isolates (0.32%) for Proteus spp.). Gram-positive bacterial isolates were 21 isolates (75%) for S. aureus and 7 isolates (25%) for E. faecalis, as shown in figure-2.
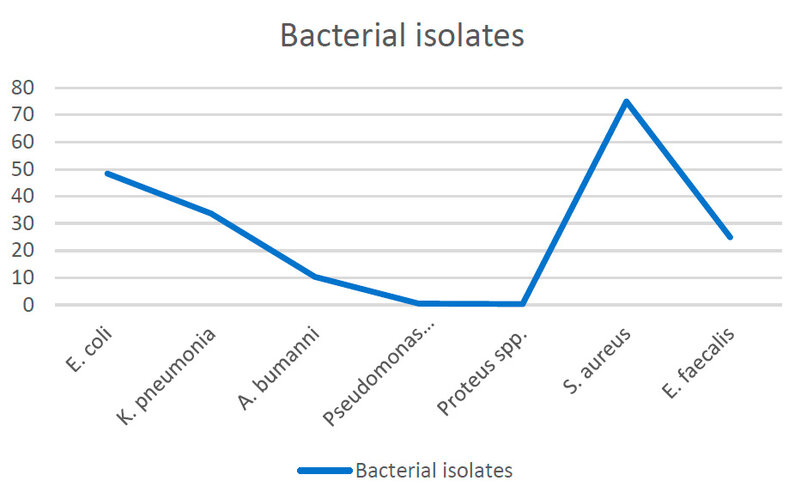
Figure 2: Bacterial isolates according to collected samples.
Antimicrobial sensitivity testing
In this study, there are 5 different antimicrobials were used. The results showed that 17 (89.47%) isolates exhibited a high resistance rate to ceftazidime, followed by cefotaxime and ceftriaxone in 15 (78.94%) isolates. On the other hand, the A. Baumann showed notably lowest resistance to imipenem and meropenem 13(68.42%) isolates and 12(63.15%), respectively, as shown in Figure 3.
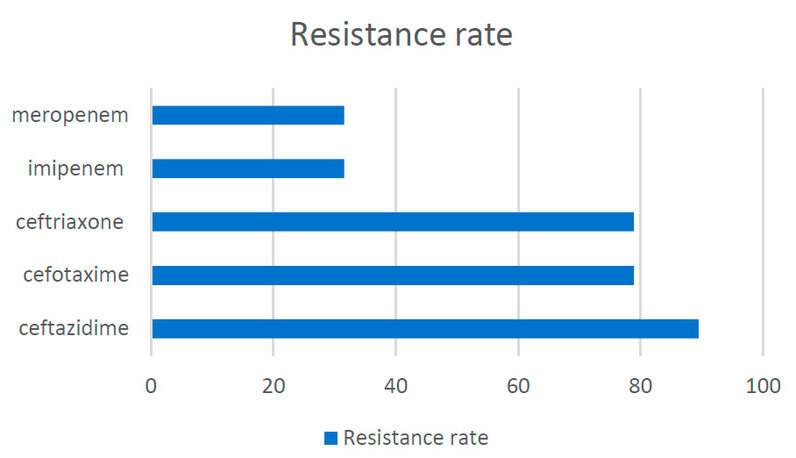
Figure 3: Antimicrobial susceptibility patterns for A. baumannii.
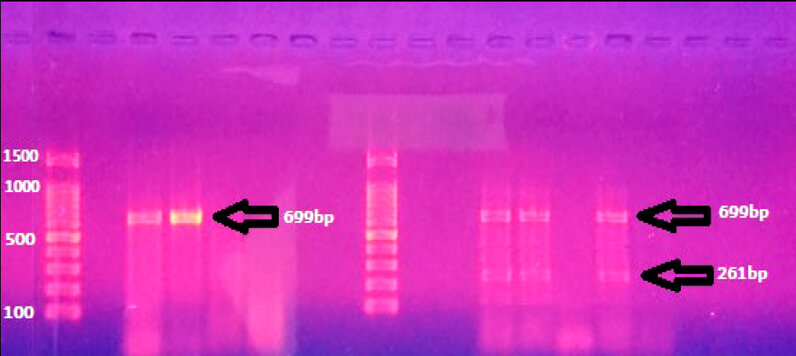
Figure 4: Gel electrophoresis of blaNDM (699bp) and blaVIM (261bp) PCR products.
Carbapenem's resistance-associated genes:
Carbapenemase-producing bacteria due to the acquisition of various antimicrobial resistance. A. baumannii causes a variety of infections, such as UTI, bacteremia, septicemia, catheter-associated infections, and wound infections. In the present study, most of the A. baumannii were recovered from urine (n=19). Similarly, a study conducted in Hong Kong also reported that 20.4% and 13% of A. baumannii were recovered from urine culture in Iran. Antimicrobial resistance has been seen from the initial use of these drugs and is a growing global concern. Acinetobacter baumannii treatment of these infections is often tricky, and carbapenems are currently the antibiotics of choice. In this study, there is an emergence of remarkable resistance to carbapenem antibiotics (imipenem and meropenem 31.58% and 36.85%, respectively); in Iran, it has been proved that the A. baumannii was isolated from UTI and showed resistance to imipenem and meropenem (78% and 44%) respectively. Also, another study in Iran by Alavi-Moghadam et al. in 2014 showed that 100% of isolates were resistant to imipenem. Carbapenemases belong to the B-lactamases classes (A, B, and D), enzymes that play the main role in B-lactam resistance. In A. baumannii bla (KPC, VIM, and NDM), genes are encoded for carbapenemases the class A (i.e., blaKPC) exhibiting a minor role in phenotypic resistance. The results indicated that 5 isolates (26.31%) were carrying the blaNDM gene, as shown in (Figure-4); this result agrees with 1, 9, and 10. Twenty percent of A. baumannii PCR results employing gene-specific primers included the blaNDM-1 gene. Furthermore, forty percent of A. baumannii isolates were found to be blaNDM-2 gene-positive when their DNA was amplified using blaNDM-2 primers. Carbapenems are a kind of -lactam antibiotic that has strong antibacterial action against Acinetobacter baumannii. However, the emergence and dissemination of acquired carbapenem resistance in these species have questioned the efficacy of treatment and control measures. In bla (NDM-1) genes, they are responsible for carbapenem resistance by encoding enzymes that partly hydrolyze carbapenems and other B-lactams 2. Only two bactericidal drugs, colistin and fosfomycin, and one bacteriostatic drug are effective against bla NDM-1 producers (tigecycline). According to the study, One of the most distinguishing features of NDM producers is that they are not only nosocomial pathogens but also Gram-negative community members, such as Acinetobacter baumannii, which can operate as a natural reservoir for bla (NDM) genes in Enterobacteriaceae 3. Thus, screening and detecting NDM-1 A. baumannii producers is necessary to aid inappropriate treatment. On the other hand, three isolates (18.75%) were carrying the blaVIM gene. The discovery of MBL-producing organisms has been difficult using phenotypic approaches such as the double-disc synergy test, MBL E-test, and combination disk test; however, molecular technologies, particularly next-generation sequencing, will throw some light on their detection 7. Phenotypic techniques only find some MBL-producing strains since they are not sensitive enough. As a result of the discovery of blaVIM-1 using PCR in 14.3% of A. baumannii isolates that had been determined to be MBL negative by E-test, it is clear how important it is to use molecular methods in everyday practice to find these concealed MBLs. Due to the lack of fitness cost and NDM lactamases being favored over other MBLs, they do not hinder bacterial development and have spread worldwide among Gram-negative bacteria. This finding conflicts with 4. Carbapenemase genes from class A, blaKPC, even though the bacterial isolates had not possessed the blaKPC gene. According to reports, Kuwait and America 5 had a 75% and 18% prevalence of bla GES, respectively. The prevalence of blaKPC in A. baumannii is rarely observed. A.baumannii clinical As this gene is found on the transportable elements, which may be passed from one bacterium to another and across the community, isolates containing the blakpc gene are disseminated in Iran. The blakpc gene's presence in These mechanisms is necessary for the formation of A.baumannii strains that are resistant to a variety of innate and acquired antimicrobials (carbapenems are the selective antibiotic for treating the infections caused by multiple-drug resistant A.baumannii strains). Therefore, the judicious use of antibiotics for infection control in hospitals, particularly in ICU departments, can significantly delay the emergence of these resistant strains and the diseases they are associated with. This amplifies the significance of prudent antibiotic use in hospitals for managing infections, especially in the intensive care unit, and lowers the risk of nosocomial transmission and potential outbreaks 6.
CONCLUSIONS
Numerous Acinetobacter baumannii isolates were found to harbor the carbapenem resistance genes, namely blaNDM and blaVIM, indicating their genetic predisposition for carbapenem resistance. Notably, the blaKPC gene was conspicuously absent in these isolates. These findings indicate a multifaceted resistance profile, as these isolates also displayed phenotypic resistance to a broad spectrum of commonly prescribed antibiotics. This highlights the complexity of antibiotic resistance mechanisms employed by Acinetobacter baumannii and raises concerns about the efficacy of conventional treatment approaches. The presence of carbapenem resistance genes underscores the clinical challenge posed by this pathogen, necessitating ongoing vigilance and Non-payment of antibiotics without laboratory tests in order to limit the spreading of bacteria resistance, especially in hospitals and create units or specialized teams in each hospital or medical center interested in developing high-definition programs for the use of antibiotics.
Author Contributions: “Conceptualization, T.H.H. and I.A.A.; methodology, T.H.H. and S.G.A.; software,A.Y.S.A.; valida-tion, S.G.A., I.A.A and A.S.Y.A.; formal analysis, I.A.A.; investigation, T.H.H..; resources, S.G.A.; data curation, I.A.A and S.G.A.; writing—original draft preparation, A.S.Y.A.; writ-ing—review and editing, I.A.A.; visualization, I.A.A.; supervision, T.H.H.; project administration, I.A.A.; funding acquisition, S.G.A., I.A.A and A.S.Y.A. All authors have read and agreed to the published version of the manuscript."
Funding: "This research received no external funding".
Informed Consent Statement: Not applicable
Acknowledgments: The authors would like to express their thanks and appreciation to the presidency of the University of Alkafeel for its support in completing this research.
Conflicts of Interest: "The authors declare no conflict of interest."
REFERENCES
1- Hasan TH, Alasedi KK, Aljanaby AA. A comparative study of prevalence antimicrobials resistance Klebsiella pneumonia among different pathogenic bacteria isolated from patients with urinary tract infection in Al-Najaf City, Iraq. Latin American journal of pharmacy. 2021 Apr 1;40:174-8
2- Al-Ethari AS, Hasan TH, Tikki KA, Bustani GS. Genotypic Detection of qnrA and qnrC Genes in Citrobacter koseri Isolated from Patients with Urinary Tract Infection. Institut Razi. Archives. 2022 Mar 1;77(2).
3-Hasan, T.H., Kadhum, H.A., Al-Khilkhali, H.J.B. Epidemiology of VZV virus in Najaf Government, Iraq. Latin American Journal of Pharmacy, 2023, 42(Special Issue), pp. 188–190.
4-Safari M, Mozaffari Nejad AS, Bahador A, Jafari R, Alikhani MY. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi J Biol Sci,2015 22: 424-29.
5-El-Shazly S, Dashti A, Vali L, Bolaris M, Ibrahim AS.Molecular epidemiology and characterization of multiple drug-resistant (MDR) clinical isolates of Acinetobacter baumannii. Int J Infect Dis,2015, 41: 42-49.
6-Hassan, L.A., Darweesh, M.F., Hasan, T.H. The Immune-modulator Activity of Pseudomonas aeruginosa Extracted Protein: Azurin Charming Protein. Latin American Journal of Pharmacy, 2023, 42(Special Issue), pp. 245–251.
7-Moulana, Z.; Babazadeh, A.; Eslamdost, Z.; Shokri, M.; Ebrahimpour, S. Phenotypic and genotypic detection of Metallo-beta-lactamases in Carbapenem-resistant Acinetobacter baumannii. Casp. J. Intern. Med,2020, 11, 171–176.
8-López, C.; Ayala, J.A.; Bonomo, R.A.; González, L.J.; Vila, A.J. Protein determinants of dissemination and host specificity of metallo-β-lactamases. Nat. Commun,2019, 10, 3617.
9-Al-Muhanna, S. G., Banoon, S. R., & Al-Kraety, I. A. A. Molecular Detection Of Integron Class 1 Gene In Proteus Mirabilis Isolated From Diabetic Foot Infections. Plant Archives, 2020, Volume 20 No.1 Pp. 3101-3107.
10-Al-Kraety, I. A.A, Al-Muhanna, S. G.& Banoon, S. R. .Molecular Exploring of Plasmid-mediated Ampc beta Lactamase Gene in Clinical Isolates of Proteus mirabilis. Revistabionatura,2021, Volume 6 No. 3 .
11-Wyres, K. L., & Holt, K. E. Klebsiella pneumoniae as a key trafficker of drug resistance genes from environmental to clinically significant bacteria. Current opinion in microbiology, 2018, 45, 131-139.
12- Aljanaby AA, Al-Faham QM, Aljanaby IA, Hasan TH. Epidemiological study of mycobacterium tuberculosis in Baghdad governorate, Iraq. Gene Reports. 2022 Mar 1;26:101467.
13-Hayder T and Aljanaby AAJ. Antibiotics susceptibility patterns of Citrobacter freundii isolated from patients with urinary tract infection in Al-Najaf governorate – Iraq. Int.J.Pharm.Sci. 2019a, 10(2): 1481-1488.
14-Hayder T and Aljanaby AAJ. Genotypic characterization of antimicrobial resistance-associated genes in Citrobacter freundii isolated from patients with urinary tract infection in Al-Najaf Governorate-Iraq. OnLine Journal of Biological Sciences,2019 b, 19 (2): 132.145.
15-Majeed, H.T., Hasan, T.H. and Aljanaby, A.A.J. Epidemiological study in women infected with toxoplasma gondii, rubella virus, and cytomegalovirus in Al-Najaf Governorate-Iraq. International Journal of Pharmaceutical Research, 2020;12, pp.1442-1447.
16-Kadhum HA, Hasan TH. The Study of Bacillus Subtils Antimicrobial Activity on Some of the Pathological Isolates. International Journal of Drug Delivery Technology,2019 21;9(02):193-6.
17-Hasan TH, Al-Harmoosh RA. Mechanisms of Antibiotics Resistance in Bacteria. Sys Rev Pharm. 2020,11(6):817-23.
18-Hasan TH. Extended Spectrum Beta Lactamase E. Coli isolated from UTI Patients in Najaf Province, Iraq. International Journal of Pharmaceutical Research.2020,12(4).
19- Bauer, R. A., & Wortzel, L. H. Doctor's choice: The physician and his sources of information about drugs. Journal of Marketing Research,1996, 3(1), 40-47.
20-Firoozeh F, Mahluji Z, Shams E, Khorshidi A, Zibaei M. New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae isolates in hospitalized patients in Kashan, Iran. Iranian journal of microbiology. 2017 Oct;9(5):283.
21- Bina M, Pournajaf A, Mirkalantari S, Talebi M, Irajian G. Detection of the Klebsiella pneumoniae carbapenemase (KPC) in K. pneumoniae Isolated from the Clinical Samples by the Phenotypic and Genotypic Methods. Iranian journal of pathology. 2015;10(3):199.
22-Abdulla, Y.N., Aljanaby, A.J.I., Hasan, H.T., Aljanaby, A.J.A. Assessment of ß-lactams and Carbapenems Antimicrobials Resistance in Klebsiella Oxytoca Isolated from Patients with Urinary Tract Infections in Najaf, Iraq.Archives of Razi Institute, 2022, 77(2), pp. 669–673
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 February 2024
Citation: Hasan T. H., AL-Ethari A. S. Y., Al-Muhanna S.G., Al-Kraety I. A. A. Genotypic Detection of Carbapenems Resistance Genes in Acinetobacter baumannii Isolated from Urinary Tract Infection Patients. Revis Bionatura 2024;1 (1) 60. http://dx.doi.org/10.21931/BJ/2024.01.01.60
Additional information Correspondence should be addressed to [email protected]
Peer review information. Bionatura thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
