2024..01.01.64
Files > Conference Series > 2024 > Chimboazo ild pagina nueva
Comparative Analysis of Threshold Cycle Results for RNA Extraction in SARS-CoV-2 RT-qPCR Using Magnetic Beads and Spin Column Methods
Fardiah Tilawati Sitanggang1*, James Perdinan Simanjuntak2*, Nasrah Nasrah3, Arvida Bar5
Ridwansyah Ridwansyah4,
1Department of Medical Laboratory Technology, Health Polytechnic of Ministry of Health Jambi, Jambi, Indonesia, [email protected],
2Department of Medical Laboratory Technology, Health Polytechnic of Ministry of Health Jambi, Jambi, Indonesia, [email protected],
3Department of Medical Laboratory Technology, Health Polytechnic of Ministry of Health Jambi, Jambi, Indonesia, [email protected],
4Nursing Department, Health Polytechnic of Ministry of Health Jambi, Jambi, Indonesia, [email protected],
5Department of Medical Laboratory Technology, Health Polytechnic of Ministry of Health Jambi, Jambi, Indonesia, [email protected],
*Correspondence: [email protected]; Tel.: +62 853-6600-3099
Available from. http://dx.doi.org/10.21931/BJ/2024.01.01.64ABSTRACT
Coronavirus Disease-2019 (COVID-19) belongs to the large family of SARS-CoV viruses, initially emerging in 2002-2003. In humans, this virus triggers respiratory infectious diseases. COVID-19, a new variant of SARS-CoV, was identified in humans following an unprecedented incident in Wuhan, China, in December 2019. This virus typically manifests mild symptoms, including a runny nose, sore throat, cough, and fever. The Nucleic Acid Amplification Test (NAAT), specifically the realtime Reverse Transcription Polymerase Chain Reaction (rRT-PCR) examination, is recommended by the World Health Organization (WHO) for diagnosing COVID-19. This study assessed potential differences in Threshold Cycle results during RNA extraction using magnetic beads compared to spin columns in the SARS-CoV-2 rRT-PCR method. The population for this study was selected through accidental sampling from nasopharyngeal and oropharyngeal swabs of COVID-19 patients obtained between December 2022 and April 2023, with Threshold Cycle values <30,000. The samples were stored at -80°C. The findings revealed that the average N (VIC) was 23.359, and RdRP (FAM) was 25.558 in the Magnetic Beads method, indicating a lower value compared to the average N (VIC) of 29.200 and RdRP (FAM) of 29.661 in the Spin Column method. This suggests that the Magnetic Beads method exhibited greater sensitivity than the Spin Column method. The statistical analysis confirmed these differences, with a P value of 0.003 in N (VIC) and the P value of 0.000 in RdRP (FAM). Consequently, it can be concluded that there is a significant 19.5% difference in the Threshold Cycle during RNA extraction using Magnetic Beads and Spin Column in the examination of the SARS-CoV-2 rRT-PCR method.
Keywords: Sars-CoV-2; rRT-PCR; Magnetic Beads; Spin Column; Threshold Cycle.
INTRODUCTION
Coronavirus Disease-2019 (COVID-19) is part of the large family of SARS-CoV viruses that initially emerged between 2002 and 2003. In humans, this virus leads to respiratory infectious diseases 1,2. This virus causes mild symptoms such as a runny nose, sore throat, cough, and fever. About 80% of cases can be recovered without specialized treatment. Patients with comorbidities and the elderly are at a higher risk of developing severe, potentially fatal diseases 3.
The Nucleic Acid Amplification Test (NAAT), specifically the realtime Reverse Transcription Polymerase Chain Reaction (rRT-PCR) examination, is recommended by the World Health Organization (WHO) for diagnosing COVID-19. The PCR technique amplifies the genetic material of the virus, SARS-CoV-2, making it a susceptible, specific, and rapid method 4,5. Before entering the PCR stage, specimen preparation is necessary, involving the extraction of nucleic acids from clinical specimens obtained from nasopharyngeal and oropharyngeal swabs 6,7.
The process of extracting DNA and RNA genetic material follows the principle of breaking cells and separating the genetic material within the cells—DNA and RNA—from other cellular components like fats, proteins, carbohydrates, and other substances. These additional components can affect the purity and integrity of the extracted genetic material. After extraction, storing the RNA base in the appropriate medium is essential to preserve its integrity and quality 8–11.
The spin column extraction method employs a silica membrane to capture DNA released from the cell. DNA-containing samples are introduced into a column containing silica gel or silica beads, along with chaotropic salts. These salts disrupt the hydrogen bonds between strands, facilitating DNA binding to silica by rendering the nucleic acid hydrophobic 12,13. The DNA binds to the silica, while the remaining solution undergoes ethanol wash to remove chaotropic salts and other extraneous elements. One limitation of this approach is its relatively extended timeframe, spanning approximately 2 to 24 hours from the initiation of cell lysis to the completion of DNA extraction 14,15.
The Magnetic Beads method employs magnet-assisted separation in nucleic acid purification. It is based on the modified alkaline lysis principle, with subsequent binding of nucleic acids to magnetic particles 16–18. A magnetic instrument captures nucleic acids bound to magnetic particles, and impurity contaminants are removed through a wash with the provided buffer. The nucleic acid is then eluted from the magnetic particles using a designated elution buffer. Despite its high effectiveness and efficiency, it may pose a challenge for small to medium-sized laboratories due to the associated costs of reagent kits and instruments 19–21.
RT-PCR serves as a technique for the amplification (multiplication) of viral nucleic acids. This amplification process is initiated by binding specific primers and probes to the target segment of the gene 22–24. The polymerase enzyme facilitates the multiplication process. In rRT-PCR, the detection of amplification products can be directly observed, eliminating the need for post-amplification stages such as gel reading or electrophoresis 7,25,26.
In 2021, the number of COVID-19 cases continued to increase by 27. This surge in cases overwhelmed the UPTD Jambi City Regional Health Laboratory due to the substantial volume of samples, reaching up to 450 per day. Currently, the Spin Column and Magnetic Beads Extraction methods are employed to examine SARS-CoV-2 at the UPTD Jambi City Regional Health Laboratory. A laboratory assistant performs The Spin Column method manually, while the Magnetic Beads method is conducted using an Automated DNA and RNA Extraction System tool. However, it remains to be seen whether there is a difference in the extraction results between these two methods 16,28,29.
Cecilia Ambrosi et al. 30 research demonstrated that nasopharyngeal and oropharyngeal swab samples, when extracted using the Spin Column method with the Qiamp DSP Virus Spin kit, exhibited higher sensitivity and maintained RNA purity. Similarly, the research by Zhen Zhao et al. 31 indicated that extraction using the Magnetic Nano Particle method, whether done manually or with automatic machines, proved more time-efficient. This method eliminated the need for toxic reagents and allowed for manual or automated (robotic) execution, ensuring high purity and productivity.
The literature review findings indicate a scarcity of studies comparing the two methods. Notably, more research must be applied to both methods to examine the SARS-CoV-2 rRT-PCR method.
Based on the findings of several studies highlighting the advantages of each RNA extraction method, the author is interested in conducting a study to determine which RNA extraction method is more effective and efficient by comparing the Threshold Cycle results of the two extraction methods.
MATERIALS AND METHODS
Study design
The type of research used in this study is analytical experimental by comparing the results of 2 types of Sars-CoV-2 RNA extraction methods by looking at the Threshold Cycle results from reading the rRT-PCR tool. The research design employs a post-test-only control group design, a specific experimental design designed to assess and compare the effectiveness of two interventions 32.
Samples
This study was conducted from March to May 2023 at the Molecular Biology Laboratory of the UPTD Health Laboratory of Jambi City. Samples were collected from nasopharyngeal and oropharyngeal swabs of COVID-19 patients from December 2022 to April 2023, with a Threshold Cycle value < 30,000. The samples were stored in a refrigerator at -80°C. The study included a sample size of 30 individuals meeting the criteria of having Acute Respiratory Infection (ARI) and, in the 14 days preceding symptom onset, having been in contact with confirmed COVID-19 patients, selected through simple randomization.
Data collection
Data were obtained from the results of the Magnetic Beads Method and the Spin Column Method by analyzing the same 30 samples with 2 different extraction methods and continued with the reading of RNA Amplification on the rRT-PCR tool.
Variables
The dependent variable is the Threshold Cycle of RNA Extraction using the Spin Column Method, measured through the realtime PCR method with the rRT-PCR measuring instrument. The independent variable is the Threshold Cycle of RNA Extraction using the Magnetic Beads Column Method, measured through the realtime PCR method with the rRT-PCR measuring instrument.
Realtime Reverse Transcription Polymerase Chain Reaction (rRT-PCR) propagates DNA templates or Complementary DNA (cDNA) using the Taq Polymerase enzyme in vitro 6.
Manual Extraction Procedure (Spin Column Method)
First, dilute the Reagent Kit, prepare the Micro Tube, VNE Column, and Elution Tube and Label the microtube to avoid swapping samples. Vortex the sample until it is homogeneous. Pipette 560 ul VNE Buffer into the Microtube, add 140 ul sample, then vortex until homogeneous, then Incubate for 10 minutes. Add 560 ul Absolute Ethanol, then vortex again; transfer into the spin column, then centrifuge for 10 seconds at 5000 X.g; prepare a vacuum Manifold, then attach the spin column to the vacuum manifold hole. Add wash buffer 1 as much as 500 ul, then suck until the liquid is wasted into the vacuum manifold, then add wash buffer 2 as much as 1000 ul, then suck until the liquid is wasted into the vacuum manifold, put it back into the Elution Tube. Centrifuge the spin column for 1 minute at 10,000 X.g to remove residual liquid. Discard the elution tube and replace it with the microtube. Add 50 ul of RNase - Freewater and incubate for 1 minute. Centrifuge again at 10,000 X.g for 1 minute. Finally, discard the spin column and store the Microtube that contains RNA, then hand it over to the clerk in the Template room.
Automated Extraction Procedure (Magnetic Beads Method)
Turn on the Automated DNA & RNA Extraction System tool. Before use, use UV first for 15 minutes. Prepare the Plate/Cartridge Extraction Kit and the Automated DNA and RNA Extraction System sample map. Homogenize the Cartridge Extraction Kit and sample using a Vortex Mixer. Add 200µ sample to each well in the 1st and 7th column series according to the sample map, and work inside the BSC. Open the glass front cover of the device. Place the plate/Cartridge in the device. Put the mixing sleeve in place. Select the desired type of examination, here selected for Sars-CoV-2 (Covid-19) examination, then press OK. After the extraction, open the Automated DNA & RNA Extraction System, then discard the Mixing Sleeve into the infectious waste container. Take the plate/cartridge and place it on the BSC. Pipette the extraction results in as much as 45µ in column 6 and column 12, and save the RNA into a microtube. Hand over the RNA to the clerk in the template room. Notes: The sample pipetting and extraction process is carried out in the biosafety cabinet
Buffer Mix Preparation Procedure (Master Mix Room)
Prepare the MasterMix Kit. Prepare plate, aluminum foil, tips, waste container, ice pack, and alcohol tissue. Put Pipette Universal Probes Reaction Mix as much as 10 ul into Microtube. Pipette Reverse Transcriptase as much as 0.2 ul into the microtube. Pipette RNase Inhibitor as much as 0.4 ul into the microtube. Pipette Primer-Probes Mix of 1.5 ul into the microtube. Pipette 2.9 ul of nuclease-free water into the microtube. Vortex the microtube until the reagents are homogeneous at 1700 RPM. Pipette 15 ul into each healthy plate (depending on the number of samples examined). Submit to the Template Note clerk: The above volume is for 1 sample, and the above process is done in Laminar flow.
Mixing Procedure of Buffer Mix and RNA (Template Chamber)
Prepare the extracted RNA. Vortex the microtube containing RNA. Pipette 5 ul of RNA into the well containing MasterMix reagent. Pipette positive control into column (12 H). Close the plate using Optical Seal, then hand it over to the PCR Application room clerk. Note: The above process is carried out in the BSC.
PCR Amplification Procedure
Turn on RT-PCR Quant Studio 5 and connect to the PC. Open the DA1 Application and input data from the MasterMix Reagent Kit into the application. Input target channels FAM (Helicase), HEX (RdRP), and Cy5 (RPP30) in the application for each well to be read. Insert the plate from the template room into the QuantStudio 5 RT-PCR tool and then run from the PC with the input data. After running is complete, open the DA2 application to read the results.
Statistical Data
Analysis of this research data to determine the comparison of CT Value results on RNA Extraction using Magnetic beads and Spin Column on Sars-CoV-2 Examination RT-PCR Method. Statistical unpaired T-tests were calculated using the SPSS version 16.0 application program. The unpaired t-test compares means between two independent groups, utilizing interval or ratio data scales.
RESULTS
The disparity in results between the Magnetic Beads method and the Spin Column method is depicted in Figure 1. Notably, the average Threshold Cycle values differ for each target, with 5.841 for target N (VIC) and 4.102 for target RdRP (FAM).
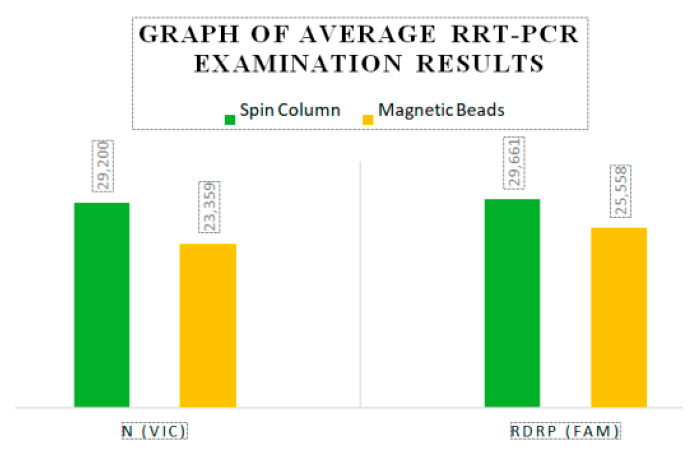
Figure 1. Average RRT-PCR examination results
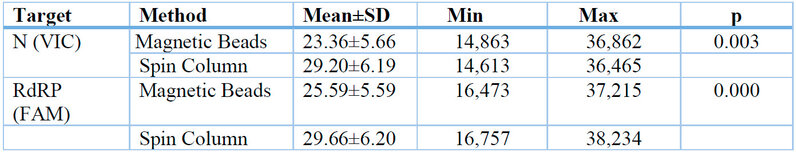
Table 1: Threshold Cycle Differences in RNA Extraction using Magnetic Beads and Spin Column in the Examination of Sars-CoV-2 rRT-PCR Method.
Table 1 shows that the average N (VIC) 23.359 and RdRP (FAM) 25.558 in the Magnetic Beads method has a lower value than the average N (VIC) 29.200 and RdRP (FAM) 29.661 in the Spin Column method, which value indicates the Magnetic Beads method is more sensitive than the Spin Column method. The results of the paired t-test showed a p-value <0.05 in N (VIC) and RdRP (FAM), so it can be concluded that there is a significant difference from the Threshold Cycle in RNA extraction using Magnetic Beads and Spin Column in the examination of Sars-CoV-2 rRT-PCR method.
DISCUSSION
This study involved 30 nasopharyngeal and oropharyngeal swab samples using Magnetic Beads and Spin Column in the examination of the rRT-PCR method for Sars-CoV-2 at the UPTD Regional Health Laboratory of Jambi City. The results of the Magnetic Beads and Spin Column extraction methods showed different Threshold Cycle values. The average Threshold Cycle for the Magnetic Beads method on the N (VIC) target was 23.359, and on the RdRP (FAM) target was 25.558, while the average Threshold Cycle for the Spin Column method on the N (VIC) target was 29.200, and on the RdRP (FAM) target was 29.660. According to the interpretation of the results using the Tianlong bufferMix Reagent kit used in this study, higher Threshold Cycle values indicate lower patient infection rates. Samples with Threshold Cycle values above 40,000 were declared as Negative.
Instruments such as micropipettes and rRT-PCR tools must be calibrated yearly to ensure testing quality 33. Based on the results of this study, the Magnetic Beads method is more efficient and effective, as it maintains RNA purity and minimizes contamination. This aligns with the findings of Zhen Z 31, which pointed out one of the drawbacks of column-based extraction: the difficulty in automation. In contrast, the Magnetic Nano Particle (MNP) method allows comprehensive nucleic acid purification, which is crucial for SARS-CoV-2 diagnosis. Overall, the improved Magnetic Beads-based extraction method exhibits high extraction efficiency and PCR amplification compatibility across various patterns, simplifying sample processing quickly and making it highly suitable for SARS-CoV-2 rRT-PCR testing.
Instruments such as micropipettes and rRT-PCR tools should undergo annual calibration to ensure the accuracy of testing33. The study's results conclude that the Magnetic Beads method is more efficient and effective, maintaining RNA purity while minimizing contamination. This finding aligns with the observations of Zhen Z 31, who highlighted a limitation of column-based extraction—precisely, the automation challenge. In contrast, the Magnetic Nano Particle (MNP) method enables comprehensive nucleic acid purification, crucial for SARS-CoV-2 diagnosis. Overall, the enhanced Magnetic Beads-based extraction method demonstrates high efficiency in extraction and compatibility with PCR amplification across various patterns. This simplification of sample processing makes it highly suitable for SARS-CoV-2 rRT-PCR testing.
According to research by Ni'mah D 34, the filter-based kit (FBK) was developed from the silica-gel approach, evolving into the silica membrane spin column method. However, a drawback of the silica membrane is its inability to effectively purify DNA in the presence of phenolic compounds and humic substances, which can bind to the membrane intended for DNA binding. Moreover, the potential for RNA contamination arises due to suboptimal RNAse handling. The Spin Column method entails numerous steps, such as replacing 4-6 microcentrifuge tubes, multiple stages of incubation, precipitation, elution, washing, and drying. This often requires specialized equipment and results in suboptimal purity. Additionally, the Spin Column method can be wasteful, necessitating changing microtubes and transferring supernatant to glass containers multiple times during extraction. In contrast, the Magnetic Beads method eliminates the need to change microtubes, contributing to its overall efficiency 17.
Sodium polyanethole sulfonate (SPS) is an anti-coagulant and an inhibitor in the PCR process. DNA, a polyanion molecule, can readily bind to the silica column. During the DNA extraction process, SPS binds to silica owing to the presence of chaotropic molecules, leading to the entrapment of both DNA and SPS in the silica column. Unfortunately, the centrifugation process proves ineffective in removing SPS from the column since the molecule is too large to pass through the membrane.
SPS, co-trapped with DNA on the column, hinders DNA escape during the elution step. SPS's large molecules effectively clog the silica membrane, impeding DNA release in the final extraction stage. Consequently, the DNA extraction results obtained through the spin column method lack DNA. Subsequent steps involving PCR and electrophoresis proved unsuccessful since the extraction results did not contain the necessary DNA.
CONCLUSIONS
The results of RNA extraction using the Magnetic Beads and Spin Column methods exhibit a notable disparity. Through statistical and average calculations, we have determined that the Magnetic Beads method is 19.5% more sensitive than the Spin Column method when extracting SARS-CoV-2 virus RNA.
Author Contributions: "Conceptualization, AV and JPS; methodology, software, validation, FTS and JPS; formal analysis, JPS; investigation, resources, data curation, NAS and RID; writing—original draft preparation, FTS; writing—review and editing, JPS.
Funding: This research received no external funding.
Institutional Review Board Statement: The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Health Research Ethics Commission, Polytechnic Health Ministry of Jambi, Indonesia, number: LB 02.06/2/531/2023
Informed Consent Statement: Patients who will undergo swabbing or potential respondents are provided with an explanation of informed consent by the researcher. The consent to participate as a respondent is conveyed by signing a statement expressing willingness.
Acknowledgments: The authors wish to extend their gratitude to the director of the Health Polytechnic, Ministry of Health, Jambi, Indonesia, for providing support during the execution of this research. Additionally, the authors would like to thank all those who contributed to this study, including the participants and the laboratory where the samples were examined.
Conflicts of Interest: The authors declare no conflict of interest.
REFERENCES
1. Hu B, Guo H, Zhou P, Shi Z-L. Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews Microbiology. 2021;19(3):141–54.
2. Kumar S, Nyodu R, Maurya VK, Saxena SK. Host immune response and immunobiology of human SARS-CoV-2 infection. Coronavirus Disease 2019 (COVID-19) Epidemiology, Pathogenesis, Diagnosis, and Therapeutics. 2020;43–53.
3. WHO U, UNDP FAO. COVID-19 UPDATE. WHO Switzerland; 2022.
4. Zhang WS, Pan J, Li F, Zhu M, Xu M, and H et al. Reverse transcription recombinase polymerase amplification coupled with CRISPR-Cas12a for facile and highly sensitive colorimetric SARS-CoV-2 detection. Analytical chemistry. 2021;93(8):4126–33.
5. Li C, Debruyne DN, Spencer J, Kapoor V, Liu LY, Zhou B, et al. High sensitivity detection of coronavirus SARS-CoV-2 using multiplex PCR and a multiplex-PCR-based metagenomic method. BioRxiv. 2020;2020:988246.
6. Agustianingsih S, Reorita R, Renny R. Optimal control for SIR Model with The Influence of Vaccination, Quarantine and Immigration factor. Jurnal Matematika, Statistika dan Komputasi. 2020;16(3):311–24.
7. Agustianingsih DP, Shaw R. Community disaster resilience using multi-hazard assessment during Covid-19: The case of Denpasar, Indonesia. Natural Hazards Research. 2023;
8. Volpato F, Lima-Morales D, Wink PL, Willig J, de-Paris F, Ashton-Prolla P, et al. Pooling of samples to optimize SARS-CoV-2 diagnosis by RT-qPCR: comparative analysis of two protocols. European Journal of Clinical Microbiology & Infectious Diseases. 2021;40:889–92.
9. Alcoba-Florez J, González-Montelongo R, Íñigo-Campos A, de Artola DG-M, Gil-Campesino H, Team TMTS, et al. Fast SARS-CoV-2 detection by RT-qPCR in preheated nasopharyngeal swab samples. International Journal of Infectious Diseases. 2020;97:66–8.
10. Miller M, Jansen M, Bisignano A, Mahoney S, Wechsberg C, Albanese N, et al. Validation of a self-administrable, saliva-based RT-qPCR test detecting SARS-CoV-2. MedRxiv. 2020;2006–20.
11. Alcoba-Florez J, Gil-Campesino H, de Artola DG-M, Díez-Gil O, Valenzuela-Fernández A, González-Montelongo R, et al. Increasing SARS-CoV-2 RT-qPCR testing capacity by sample pooling. International Journal of Infectious Diseases. 2021;103:19–22.
12. Bruce EA, Huang M-L, Perchetti GA, Tighe S, Laaguiby P, Hoffman JJ, et al. Direct RT-qPCR detection of SARS-CoV-2 RNA from patient nasopharyngeal swabs without an RNA extraction step. PLoS Biology. 2020;18(10):e3000896.
13. Borghei Y-S, Samadikhah HR, Hosseinkhani S. Exploitation of N-gene of SARS-CoV-2 to develop a new rapid assay by ASOs@ AuNPs. Analytical Chemistry. 2022;94(39):13616–22.
14. Graham TGW, Dugast-Darzacq C, Dailey GM, Nguyenla XH, Van Dis E, Esbin MN, et al. Open-source RNA extraction and RT-qPCR methods for SARS-CoV-2 detection. PloS one. 2021;16(2):e0246647.
15. Hirotsu Y, Maejima M, Shibusawa M, Nagakubo Y, Hosaka K, Amemiya K, et al. Pooling RT-qPCR testing for SARS-CoV-2 in 1000 individuals of healthy and infection-suspected patients. Scientific reports. 2020;10(1):18899.
16. Klein S, Müller TG, Khalid D, Sonntag-Buck V, Heuser A-M, Glass B, et al. SARS-CoV-2 RNA extraction using magnetic beads for rapid large-scale testing by RT-qPCR and RT-LAMP. Viruses. 2020;12(8):863.
17. Babler KM, Amirali A, Sharkey ME, Williams SL, Boone MM, Cosculluela GA, et al. Comparison of electronegative filtration to magnetic bead-based concentration and V2G-qPCR to RT-qPCR for quantifying viral SARS-CoV-2 RNA from wastewater. ACS ES&T Water. 2022;2(11):2004–13.
18. Parra-Guardado AL, Sweeney CL, Hayes EK, Trueman BF, Huang Y, Jamieson RC, et al. Development of a rapid pre-concentration protocol and a magnetic beads-based RNA extraction method for SARS-CoV-2 detection in raw municipal wastewater. Environmental Science: Water Research & Technology. 2022;8(1):47–61.
19. Tohari TR, Anshori I, Baroroh U, Nugroho AE, Gumilar G, Kusumawardani S, et al. Development of a Single-Chain Variable Fragment of CR3022 for a Plasmonic-Based Biosensor Targeting the SARS-CoV-2 Spike Protein. Biosensors. 2022;12(12):1133.
20. Litanto V, Kim Y, Sungono V, Siswadi Y, Nugroho EH, Jo J. Assessment on COVID-19 Antibody and Antigen Rapid Test Devices as Screening Tools for SARS-CoV-2 Infection at the Academic Premises. GCISTEM Proceeding. 2022;1:152–9.
21. Koentjoro MP, Shamsudin SB, Bermawi B, Prayekti E, Prasetyo EN. RNA Internal Control (IC) for Routine Clinical Diagnostic Realtime Reverse Transcription-PCR SARS-CoV-2. In: First International Conference on Medical Technology (ICoMTech 2021). Atlantis Press; 2022. p. 86–92.
22. Sakai J, Tarumoto N, Orihara Y, Kawamura R, Kodana M, Matsuzaki N, et al. Evaluation of a high-speed but low-throughput RT-qPCR system for detection of SARS-CoV-2. Journal of Hospital Infection. 2020;105(4):615–8.
23. Kudo E, Israelow B, Vogels CBF, Lu P, Wyllie AL, Tokuyama M, et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biology. 2020;18(10):e3000867.
24. Minami K, Masutani R, Suzuki Y, Kubota M, Osaka N, Nakanishi T, et al. Evaluation of SARS-CoV-2 RNA quantification by RT-LAMP compared to RT-qPCR. Journal of Infection and Chemotherapy. 2021;27(7):1068–71.
25. Boni MF, Lemey P, Jiang X, Lam TT-Y, Perry BW, Castoe TA, et al. Evolutionary origins of the SARS-CoV-2 sarbecovirus lineage responsible for the COVID-19 pandemic. Nature microbiology. 2020;5(11):1408–17.
26. Pavan M, Bassani D, Sturlese M, Moro S. From the Wuhan-Hu-1 strain to the XD and XE variants: is targeting the SARS-CoV-2 spike protein still a pharmaceutically relevant option against COVID-19? Journal of Enzyme Inhibition and Medicinal Chemistry. 2022;37(1):1704–14.
27. Skevaki C, Karsonova A, Karaulov A, Xie M, Renz H. Asthma-associated risk for COVID-19 development. Journal of Allergy and Clinical Immunology. 2020;146(6):1295–301.
28. Li S, Han B, Zhou D, Gu Y, Li B, Ma J, et al. One-Stop Extraction and In Situ RT-qPCR for Ultrasensitive Detection of Highly Diluted SARS-CoV-2 in Large-Volume Samples from Aquatic Environments. Analytical Chemistry. 2023;95(4):2339–47.
29. Pérez-Cataluña A, Cuevas-Ferrando E, Randazzo W, Falco I, Allende A, Sanchez G. Comparing analytical methods to detect SARS-CoV-2 in wastewater. Science of the Total Environment. 2021;758:143870.
30. Ambrosi C, Prezioso C, Checconi P, Scribano D, Sarshar M, Capannari M, et al. SARS-CoV-2: Comparative analysis of different RNA extraction methods. Journal of virological methods. 2021;287:114008.
31. Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. BioRxiv. 2020;2002–20.
32. Arifin Z. Metodologi penelitian pendidikan. Vol. 1, Jurnal Al-Hikmah. 2020.
33. Gogoi S, Bora I, Debnath E, Sarkar S, Jais MB, Sharma A. Perplexity vs Clarity in choosing the right molecular diagnostic techniques for SARS-COV2 detection in Indian setup. Journal of Family Medicine and Primary Care. 2021;10(2):615.
34. Sinaga LP, Kartika D, Ni'mah N, Monalisa C. Stability and optimal control of the coronavirus (SARS-CoV-2) SEIR model in Indonesia. In: AIP Conference Proceedings. AIP Publishing; 2022.
Received: October 9th 2023/ Accepted: January 15th 2024 / Published:15 February 2024
Citation: F. T. Sitanggang, Simanjuntak J. P., Nasrazuhdy, Ridwansyah, Bar A. Comparative Analysis of Threshold Cycle Results for RNA Extraction in SARS-CoV-2 RT-qPCR Using Magnetic Beads and Spin Column Methods. Revis Bionatura 2024; 1 (1) 64. http://dx.doi.org/10.21931/BJ/2024.01.01.64
Additional information Correspondence should be addressed to [email protected]
Peer review information. Bionatura thanks anonymous reviewer(s) for their contribution to the peer review of this work using https://reviewerlocator.webofscience.com/
All articles published by Bionatura Journal are made freely and permanently accessible online immediately upon publication, without subscription charges or registration barriers.
Publisher's Note: Bionatura stays neutral concerning jurisdictional claims in published maps and institutional affiliations.
Copyright: © 2024 by the authors. They were submitted for possible open-access publication under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
